2QS1
 
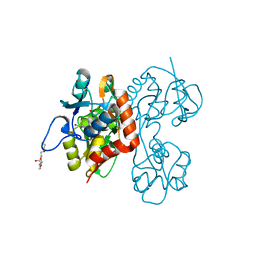 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP315 at 1.80 Angstroms resolution | | 分子名称: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-methyl-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)-4,5-dibromothiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | 著者 | Alushin, G.M, Jane, D.E, Mayer, M.L. | | 登録日 | 2007-07-30 | | 公開日 | 2008-08-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
2QS2
 
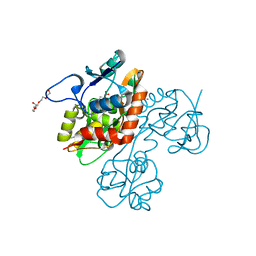 | | Crystal structure of the GluR5 ligand binding core dimer in complex with UBP318 at 1.80 Angstroms resolution | | 分子名称: | 3-({3-[(2S)-2-amino-2-carboxyethyl]-5-bromo-2,6-dioxo-3,6-dihydropyrimidin-1(2H)-yl}methyl)thiophene-2-carboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | 著者 | Alushin, G.M, Jane, D.E, Mayer, M.L. | | 登録日 | 2007-07-30 | | 公開日 | 2008-08-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
2QS4
 
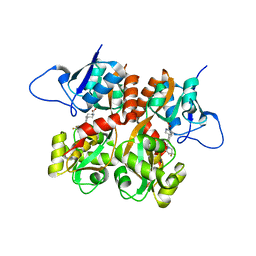 | | Crystal structure of the GluR5 ligand binding core dimer in complex with LY466195 at 1.58 Angstroms resolution | | 分子名称: | (3S,4aR,6S,8aR)-6-{[(2S)-2-carboxy-4,4-difluoropyrrolidin-1-yl]methyl}decahydroisoquinoline-3-carboxylic acid, AMMONIUM ION, GLYCEROL, ... | | 著者 | Alushin, G.M, Jane, D.E, Mayer, M.L. | | 登録日 | 2007-07-30 | | 公開日 | 2008-08-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.58 Å) | | 主引用文献 | Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists.
Neuropharmacology, 60, 2011
|
|
