1PQO
 
 | | T4 Lysozyme Core Repacking Mutant L118I/TA | | 分子名称: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Lysozyme, ... | | 著者 | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | 登録日 | 2003-06-18 | | 公開日 | 2003-10-07 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Repacking the Core of T4 lysozyme by automated design
J.Mol.Biol., 332, 2003
|
|
1PQP
 
 | | Crystal Structure of the C136S Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae Bound with Aspartate Semialdehyde and Phosphate | | 分子名称: | Aspartate-semialdehyde dehydrogenase, L-HOMOSERINE, PHOSPHATE ION | | 著者 | Blanco, J, Moore, R.A, Faehnle, C.R, Viola, R.E. | | 登録日 | 2003-06-18 | | 公開日 | 2004-08-10 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PQQ
 
 | | NMR Structure of a Cyclic Polyamide-DNA Complex | | 分子名称: | 45-(3-AMINOPROPYL)-5,11,22,28,34-PENTAMETHYL-3,9,15,20,26,32,38,43-OCTAOXO-2,5,8,14,19,22,25,28,31,34,37,42,45,48-TETRADECAAZA-11-AZONIAHEPTACYCLO[42.2.1.1~4,7~.1~10,13~.1~21,24~.1~27,30~.1~33,36~]DOPENTACONTA-1(46),4(52),6,10(51),12,21(50),23,27(49),29,33(48),35,44(47)-DODECAENE, 5'-D(*CP*GP*CP*TP*AP*AP*CP*AP*GP*GP*C)-3', 5'-D(*GP*CP*CP*TP*GP*TP*TP*AP*GP*CP*G)-3' | | 著者 | Zhang, Q, Dwyer, T.J, Tsui, V, Case, D.A, Cho, J, Dervan, P.B, Wemmer, D.E. | | 登録日 | 2003-06-18 | | 公開日 | 2004-06-29 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR Structure of a Cyclic Polyamide-DNA Complex.
J.Am.Chem.Soc., 126, 2004
|
|
1PQR
 
 | | Solution Conformation of alphaA-Conotoxin EIVA | | 分子名称: | Alpha-A-conotoxin EIVA | | 著者 | Chi, S.-W, Park, K.-H, Suk, J.-E, Olivera, B.M, McIntosh, J.M, Han, K.-H. | | 登録日 | 2003-06-18 | | 公開日 | 2003-11-04 | | 最終更新日 | 2022-03-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Conformation of alphaA-conotoxin EIVA, a Potent Neuromuscular Nicotinic Acetylcholine Receptor Antagonist from Conus ermineus
J.Biol.Chem., 278, 2003
|
|
1PQS
 
 | | Solution structure of the C-terminal OPCA domain of yCdc24p | | 分子名称: | Cell division control protein 24 | | 著者 | Leitner, D, Wahl, M, Labudde, D, Diehl, A, Schmieder, P, Pires, J.R, Fossi, M, Leidert, M, Krause, G, Oschkinat, H. | | 登録日 | 2003-06-19 | | 公開日 | 2003-07-01 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of an N-terminally truncated version of the yeast CDC24p PB1 domain shows a different beta-sheet topology.
Febs Lett., 579, 2005
|
|
1PQT
 
 | |
1PQU
 
 | | Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae Bound with NADP, S-methyl cysteine sulfoxide and cacodylate | | 分子名称: | Aspartate-semialdehyde dehydrogenase, CACODYLATE ION, CYSTEINE, ... | | 著者 | Blanco, J, Moore, R.A, Viola, R.E. | | 登録日 | 2003-06-19 | | 公開日 | 2004-08-10 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PQV
 
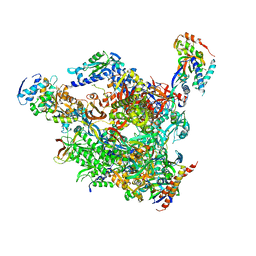 | | RNA polymerase II-TFIIS complex | | 分子名称: | DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 14.2 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, ... | | 著者 | Kettenberger, H, Armache, K.-J, Cramer, P. | | 登録日 | 2003-06-19 | | 公開日 | 2003-08-19 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Architecture of the RNA Polymerase II-TFIIS Complex and Implications for mRNA Cleavage
Cell(Cambridge,Mass.), 114, 2003
|
|
1PQW
 
 | |
1PQX
 
 | | Solution NMR Structure of Staphylococcus aureus protein SAV1430. Northeast Structural Genomics Consortium Target ZR18. | | 分子名称: | conserved hypothetical protein | | 著者 | Baran, M.C, Aramini, J.M, Xiao, R, Huang, Y.J, Acton, T.B, Shih, L, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | 登録日 | 2003-06-19 | | 公開日 | 2004-09-07 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution Strucutre of the Hypothetical Staphylococcus Aureus protein SAV1430. Northest Strucutral Genomics Consortium target ZR18
To be Published
|
|
1PQY
 
 | | Crystal structure of formyl-coA transferase yfdW from E. coli | | 分子名称: | Hypothetical protein yfdW | | 著者 | Gogos, A, Gorman, J, Shapiro, L, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | 登録日 | 2003-06-19 | | 公開日 | 2003-09-30 | | 最終更新日 | 2021-02-03 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Structure of Escherichia coli YfdW, a type III CoA transferase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PQZ
 
 | |
1PR0
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with Inosine and Phosphate/Sulfate | | 分子名称: | INOSINE, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | 著者 | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | 登録日 | 2003-06-19 | | 公開日 | 2003-11-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PR1
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with Formycin B and Phosphate/Sulfate | | 分子名称: | FORMYCIN B, PHOSPHATE ION, Purine nucleoside phosphorylase | | 著者 | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | 登録日 | 2003-06-19 | | 公開日 | 2003-11-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PR2
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 9-beta-D-[2-deoxyribofuranosyl]-6-methylpurine and Phosphate/Sulfate | | 分子名称: | 9-(2-DEOXY-BETA-D-RIBOFURANOSYL)-6-METHYLPURINE, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | 著者 | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | 登録日 | 2003-06-19 | | 公開日 | 2003-11-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PR3
 
 | | Crystal Structure of the R103K Mutant of Aspartate Semialdehyde dehydrogenase from Haemophilus influenzae | | 分子名称: | Aspartate semialdehyde dehydrogenase, PHOSPHATE ION | | 著者 | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | 登録日 | 2003-06-19 | | 公開日 | 2004-07-27 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PR4
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 9-beta-D-ribofuranosyl-6-methylthiopurine and Phosphate/Sulfate | | 分子名称: | 2-HYDROXYMETHYL-5-(6-METHYLSULFANYL-PURIN-9-YL)-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | 著者 | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | 登録日 | 2003-06-19 | | 公開日 | 2003-11-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PR5
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 7-deazaadenosine and Phosphate/Sulfate | | 分子名称: | '2-(4-AMINO-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | 著者 | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | 登録日 | 2003-06-19 | | 公開日 | 2003-11-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PR6
 
 | | Escherichia coli Purine Nucleoside Phosphorylase Complexed with 9-beta-D-xylofuranosyladenine and Phosphate/Sulfate | | 分子名称: | 2-(6-AMINO-OCTAHYDRO-PURIN-9-YL)-5-HYDROXYMETHYL-TETRAHYDRO-FURAN-3,4-DIOL, PHOSPHATE ION, Purine nucleoside phosphorylase DeoD-type | | 著者 | Bennett, E.M, Li, C, Allan, P.W, Parker, W.B, Ealick, S.E. | | 登録日 | 2003-06-19 | | 公開日 | 2003-11-25 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural basis for substrate specificity of Escherichia coli purine nucleoside phosphorylase.
J.Biol.Chem., 278, 2003
|
|
1PR9
 
 | | Human L-Xylulose Reductase Holoenzyme | | 分子名称: | DIHYDROGENPHOSPHATE ION, L-XYLULOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | El-Kabbani, O, Ishikura, S, Darmanin, C, Carbone, V, Chung, R.P.-T, Usami, N, Hara, A. | | 登録日 | 2003-06-20 | | 公開日 | 2004-02-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.96 Å) | | 主引用文献 | Crystal structure of human L-xylulose reductase holoenzyme: probing the role of Asn107 with site-directed mutagenesis
Proteins, 55, 2004
|
|
1PRA
 
 | |
1PRB
 
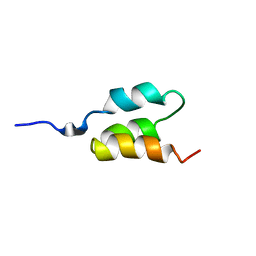 | | STRUCTURE OF AN ALBUMIN-BINDING DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | 分子名称: | PROTEIN PAB | | 著者 | Johansson, M.U, De Chateau, M, Wikstrom, M, Forsen, S, Drakenberg, T, Bjorck, L. | | 登録日 | 1997-01-15 | | 公開日 | 1997-07-23 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the albumin-binding GA module: a versatile bacterial protein domain.
J.Mol.Biol., 266, 1997
|
|
1PRC
 
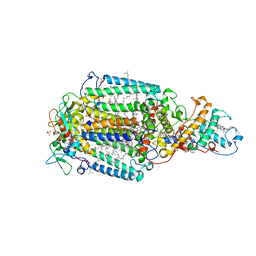 | | CRYSTALLOGRAPHIC REFINEMENT AT 2.3 ANGSTROMS RESOLUTION AND REFINED MODEL OF THE PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS | | 分子名称: | 15-trans-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | 著者 | Deisenhofer, J, Epp, O, Miki, K, Huber, R, Michel, H. | | 登録日 | 1988-02-04 | | 公開日 | 1989-01-09 | | 最終更新日 | 2021-03-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystallographic refinement at 2.3 A resolution and refined model of the photosynthetic reaction centre from Rhodopseudomonas viridis.
J.Mol.Biol., 246, 1995
|
|
1PRE
 
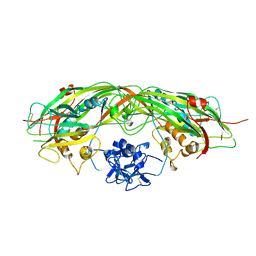 | | PROAEROLYSIN | | 分子名称: | PROAEROLYSIN | | 著者 | Parker, M.W, Buckley, J.T, Postma, J.P.M, Tucker, A.D, Tsernoglou, D. | | 登録日 | 1995-09-15 | | 公開日 | 1996-10-14 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of the Aeromonas toxin proaerolysin in its water-soluble and membrane-channel states.
Nature, 367, 1994
|
|
1PRG
 
 | |
