1V75
 
 | | Crystal structure of hemoglobin D from the Aldabra giant tortoise (Geochelone gigantea) at 2.0 A resolution | | Descriptor: | Hemoglobin A and D beta chain, Hemoglobin D alpha chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kuwada, T, Hasegawa, T, Satoh, I, Ishikawa, K, Shishikura, F. | | Deposit date: | 2003-12-12 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystallization and preliminary X-ray diffraction study of hemoglobin D from the Aldabra giant tortoise, Geochelone gigantea.
Protein Pept.Lett., 10, 2003
|
|
1V76
 
 | |
1V77
 
 | | Crystal structure of the PH1877 protein | | Descriptor: | hypothetical protein PH1877 | | Authors: | Takagi, H, Numata, T, Kakuta, Y, Kimura, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the ribonuclease P protein Ph1877p from hyperthermophilic archaeon Pyrococcus horikoshii OT3
Biochem.Biophys.Res.Commun., 319, 2004
|
|
1V79
 
 | | Crystal structures of adenosine deaminase complexed with potent inhibitors | | Descriptor: | 1-{(1R,2S)-1-[2-(2,3,-DICHLOROPHENYL)ETHYL]-2-HYDROXYPROPYL}-1H-IMIDAZOLE-4-CARBOXAMIDE, Adenosine deaminase, ZINC ION | | Authors: | Kinoshita, T. | | Deposit date: | 2003-12-14 | | Release date: | 2004-12-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design and synthesis of non-nucleoside, potent, and orally bioavailable adenosine deaminase inhibitors
J.Med.Chem., 47, 2004
|
|
1V7A
 
 | | Crystal structures of adenosine deaminase complexed with potent inhibitors | | Descriptor: | 1-{(1R,2S)-2-HYDROXY-1-[2-(2-NAPHTHYLOXY)ETHYL]PROPYL}-1H-IMIDAZONE-4-CARBOXAMIDE, ZINC ION, adenosine deaminase | | Authors: | Kinoshita, T. | | Deposit date: | 2003-12-14 | | Release date: | 2004-12-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-based design and synthesis of non-nucleoside, potent, and orally bioavailable adenosine deaminase inhibitors
J.Med.Chem., 47, 2004
|
|
1V7C
 
 | |
1V7F
 
 | | Solution structure of phrixotoxin 1 | | Descriptor: | Phrixotoxin 1 | | Authors: | Chagot, B, Escoubas, P, Villegas, E, Bernard, C, Ferrat, G, Corzo, G, Lazdunski, M, Darbon, H. | | Deposit date: | 2003-12-16 | | Release date: | 2004-11-23 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Phrixotoxin 1, a specific peptide inhibitor of Kv4 potassium channels from the venom of the theraphosid spider Phrixotrichus auratus
Protein Sci., 13, 2004
|
|
1V7H
 
 | | Crystal Structures of Collagen Model Peptides with Pro-Hyp-Gly Sequence at 1.26 A | | Descriptor: | Collagen like peptide | | Authors: | Okuyama, K, Hongo, C, Fukushima, R, Wu, G, Narita, H, Noguchi, K, Tanaka, Y, Nishino, N. | | Deposit date: | 2003-12-17 | | Release date: | 2004-08-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of collagen model peptides with Pro-Hyp-Gly repeating sequence at 1.26 A resolution: implications for proline ring puckering
Biopolymers, 76, 2004
|
|
1V7L
 
 | |
1V7M
 
 | | Human Thrombopoietin Functional Domain Complexed To Neutralizing Antibody TN1 Fab | | Descriptor: | Monoclonal TN1 Fab Heavy Chain, Monoclonal TN1 Fab Light Chain, Thrombopoietin | | Authors: | Feese, M.D, Tamada, T, Kato, Y, Maeda, Y, Hirose, M, Matsukura, Y, Shigematsu, H, Kato, T, Miyazaki, H, Kuroki, R. | | Deposit date: | 2003-12-18 | | Release date: | 2004-03-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of the receptor-binding domain of human thrombopoietin determined by complexation with a neutralizing antibody fragment
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1V7N
 
 | | Human Thrombopoietin Functional Domain Complexed To Neutralizing Antibody TN1 Fab | | Descriptor: | Monoclonal TN1 Fab Heavy Chain, Monoclonal TN1 Fab Light Chain, Thrombopoietin | | Authors: | Feese, M.D, Tamada, T, Kato, Y, Maeda, Y, Hirose, M, Matsukura, Y, Shigematsu, H, Kato, T, Miyazaki, H, Kuroki, R. | | Deposit date: | 2003-12-18 | | Release date: | 2004-03-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the receptor-binding domain of human thrombopoietin determined by complexation with a neutralizing antibody fragment
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1V7O
 
 | |
1V7P
 
 | | Structure of EMS16-alpha2-I domain complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, EMS16 A chain, ... | | Authors: | Horii, K, Okuda, D, Morita, T, Mizuno, H. | | Deposit date: | 2003-12-19 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of EMS16 in complex with the integrin alpha2-I domain
J.Mol.Biol., 341, 2004
|
|
1V7Q
 
 | |
1V7R
 
 | |
1V7S
 
 | |
1V7T
 
 | |
1V7U
 
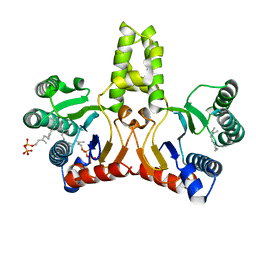 | | Crystal structure of Undecaprenyl Pyrophosphate Synthase with farnesyl pyrophosphate | | Descriptor: | FARNESYL DIPHOSPHATE, Undecaprenyl pyrophosphate synthetase | | Authors: | Chang, S.-Y, Ko, T.-P, Chen, A.P.-C, Wang, A.H.-J, Liang, P.-H. | | Deposit date: | 2003-12-24 | | Release date: | 2004-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Substrate binding mode and reaction mechanism of undecaprenyl pyrophosphate synthase deduced from crystallographic studies
Protein Sci., 13, 2004
|
|
1V7V
 
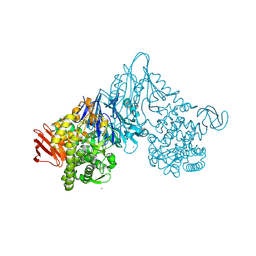 | | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase | | Descriptor: | CALCIUM ION, chitobiose phosphorylase | | Authors: | Hidaka, M, Honda, Y, Nirasawa, S, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Structure, 12, 2004
|
|
1V7W
 
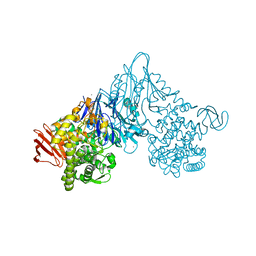 | | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Hidaka, M, Honda, Y, Nirasawa, S, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Structure, 12, 2004
|
|
1V7X
 
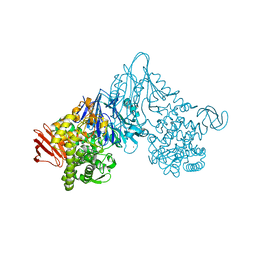 | | Crystal structure of Vibrio proteolyticus chitobiose phosphorylase in complex with GlcNAc and sulfate | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Hidaka, M, Honda, Y, Nirasawa, S, Kitaoka, M, Hayashi, K, Wakagi, T, Shoun, H, Fushinobu, S. | | Deposit date: | 2003-12-24 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Chitobiose phosphorylase from Vibrio proteolyticus, a member of glycosyl transferase family 36, has a clan GH-L-like (alpha/alpha)(6) barrel fold.
Structure, 12, 2004
|
|
1V7Y
 
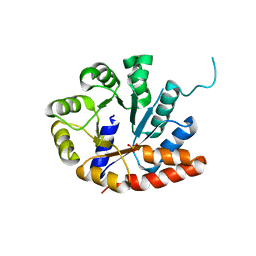 | | Crystal structure of tryptophan synthase alpha-subunit from Escherichia coli at room temperature | | Descriptor: | SULFATE ION, Tryptophan synthase alpha chain | | Authors: | Nishio, K, Morimoto, Y, Ishizuka, M, Ogasahara, K, Yutani, K, Tsukihara, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-12-25 | | Release date: | 2005-02-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conformational Changes in the alpha-Subunit Coupled to Binding of the beta(2)-Subunit of Tryptophan Synthase from Escherichia coli: Crystal Structure of the Tryptophan Synthase alpha-Subunit Alon
Biochemistry, 44, 2005
|
|
1V7Z
 
 | | creatininase-product complex | | Descriptor: | MANGANESE (II) ION, N-[(E)-AMINO(IMINO)METHYL]-N-METHYLGLYCINE, SULFATE ION, ... | | Authors: | Yoshimoto, T, Tanaka, N, Kanada, N, Inoue, T, Nakajima, Y, Haratake, M, Nakamura, K.T, Xu, Y, Ito, K. | | Deposit date: | 2003-12-26 | | Release date: | 2004-01-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of creatininase reveal the substrate binding site and provide an insight into the catalytic mechanism
J.Mol.Biol., 337, 2004
|
|
1V80
 
 | |
1V81
 
 | |
