1U1X
 
 | | Structure and function of phenazine-biosynthesis protein PhzF from Pseudomonas fluorescens 2-79 | | Descriptor: | (2S,3S)-TRANS-2,3-DIHYDRO-3-HYDROXYANTHRANILIC ACID, Phenazine biosynthesis protein phzF | | Authors: | Blankenfeldt, W, Kuzin, A.P, Skarina, T, Korniyenko, Y, Tong, L, Bayer, P, Janning, P, Thomashow, L.S, Mavrodi, D.V. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure and function of the phenazine biosynthetic protein PhzF from Pseudomonas fluorescens.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1U1Y
 
 | | Crystal structure of a complex between WT bacteriophage MS2 coat protein and an F5 aptamer RNA stemloop with 2aminopurine substituted at the-10 position | | Descriptor: | 5'-R(*CP*CP*GP*GP*(2PR)P*GP*GP*AP*UP*CP*AP*CP*CP*AP*CP*GP*G)-3', Coat protein | | Authors: | Horn, W.T, Convery, M.A, Stonehouse, N.J, Adams, C.J, Liljas, L, Phillips, S.E, Stockley, P.G. | | Deposit date: | 2004-07-16 | | Release date: | 2004-12-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The crystal structure of a high affinity RNA stem-loop complexed with the bacteriophage MS2 capsid: further challenges in the modeling of ligand-RNA interactions.
Rna, 10, 2004
|
|
1U1Z
 
 | | The Structure of (3R)-hydroxyacyl-ACP dehydratase (FabZ) | | Descriptor: | (3R)-hydroxymyristoyl-[acyl carrier protein] dehydratase, SULFATE ION | | Authors: | Kimber, M.S, Martin, F, Lu, Y, Houston, S, Vedadi, M, Dharamsi, A, Fiebig, K.M, Schmid, M, Rock, C.O. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of (3R)-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Pseudomonas aeruginosa
J.Biol.Chem., 279, 2004
|
|
1U20
 
 | |
1U21
 
 | | transthyretin with tethered inhibitor on one monomer. | | Descriptor: | 2-[(3,5-DICHLORO-4-TRIOXIDANYLPHENYL)AMINO]BENZOIC ACID, Transthyretin | | Authors: | Wiseman, R.L, Johnson, S.M, Kelker, M.S, Foss, T, Wilson, I.A, Kelly, J.W. | | Deposit date: | 2004-07-16 | | Release date: | 2005-06-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Kinetic stabilization of an oligomeric protein by a single ligand binding event
J.Am.Chem.Soc., 127, 2005
|
|
1U22
 
 | | A. thaliana cobalamine independent methionine synthase | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, 2-AMINO-4-MERCAPTO-BUTYRIC ACID, 5-methyltetrahydropteroyltriglutamate--homocysteine methyltransferase, ... | | Authors: | Ferrer, J.-L, Ravanel, S, Robert, M, Dumas, R. | | Deposit date: | 2004-07-16 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of cobalamin-independent methionine synthase complexed with zinc, homocysteine, and methyltetrahydrofolate
J.Biol.Chem., 279, 2004
|
|
1U24
 
 | | Crystal structure of Selenomonas ruminantium phytase | | Descriptor: | myo-inositol hexaphosphate phosphohydrolase | | Authors: | Chu, H.M, Guo, R.T, Lin, T.W, Chou, C.C, Shr, H.L, Lai, H.L, Tang, T.Y, Cheng, K.J, Selinger, B.L, Wang, A.H.-J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Selenomonas ruminantium Phytase in Complex with Persulfated Phytate; DSP Phytase Fold and Mechanism for Sequential Substrate Hydrolysis
STRUCTURE, 12, 2004
|
|
1U25
 
 | | Crystal structure of Selenomonas ruminantium phytase complexed with persulfated phytate in the C2221 crystal form | | Descriptor: | D-MYO-INOSITOL-HEXASULPHATE, myo-inositol hexaphosphate phosphohydrolase | | Authors: | Chu, H.M, Guo, R.T, Lin, T.W, Chou, C.C, Shr, H.L, Lai, H.L, Tang, T.Y, Cheng, K.J, Selinger, B.L, Wang, A.H.-J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Selenomonas ruminantium Phytase in Complex with Persulfated Phytate; DSP Phytase Fold and Mechanism for Sequential Substrate Hydrolysis
STRUCTURE, 12, 2004
|
|
1U26
 
 | | Crystal structure of Selenomonas ruminantium phytase complexed with persulfated phytate | | Descriptor: | D-MYO-INOSITOL-HEXASULPHATE, myo-inositol hexaphosphate phosphohydrolase | | Authors: | Chu, H.M, Guo, R.T, Lin, T.W, Chou, C.C, Shr, H.L, Lai, H.L, Tang, T.Y, Cheng, K.J, Selinger, B.L, Wang, A.H.-J. | | Deposit date: | 2004-07-16 | | Release date: | 2004-11-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Selenomonas ruminantium Phytase in Complex with Persulfated Phytate; DSP Phytase Fold and Mechanism for Sequential Substrate Hydrolysis
STRUCTURE, 12, 2004
|
|
1U27
 
 | | Triglycine variant of the ARNO Pleckstrin Homology Domain in complex with Ins(1,3,4,5)P4 | | Descriptor: | Cytohesin 2, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE | | Authors: | Cronin, T.C, DiNitto, J.P, Czech, M.P, Lambright, D.G. | | Deposit date: | 2004-07-16 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural determinants of phosphoinositide selectivity in splice variants of Grp1 family PH domains
Embo J., 23, 2004
|
|
1U28
 
 | | R. rubrum transhydrogenase asymmetric complex (dI.NAD+)2(dIII.NADP+)1 | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Mather, O.C, Singh, A, van Boxel, G.I, White, S.A, Jackson, J.B. | | Deposit date: | 2004-07-16 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Active-site conformational changes associated with hydride transfer in proton-translocating transhydrogenase.
Biochemistry, 43, 2004
|
|
1U29
 
 | | Triglycine variant of the ARNO Pleckstrin Homology Domain in complex with Ins(1,4,5)P3 | | Descriptor: | Cytohesin 2, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE | | Authors: | Cronin, T.C, DiNitto, J.P, Czech, M.P, Lambright, D.G. | | Deposit date: | 2004-07-16 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants of phosphoinositide selectivity in splice variants of Grp1 family PH domains
Embo J., 23, 2004
|
|
1U2A
 
 | |
1U2B
 
 | | Triglycine variant of the Grp1 Pleckstrin Homology Domain unliganded | | Descriptor: | Cytohesin 3, SULFATE ION | | Authors: | Cronin, T.C, DiNitto, J.P, Czech, M.P, Lambright, D.G. | | Deposit date: | 2004-07-16 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural determinants of phosphoinositide selectivity in splice variants of Grp1 family PH domains
Embo J., 23, 2004
|
|
1U2C
 
 | |
1U2D
 
 | | Structre of transhydrogenaes (dI.NADH)2(dIII.NADPH)1 asymmetric complex | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, NAD(P) transhydrogenase subunit beta, ... | | Authors: | Mather, O.C, Singh, A, van Boxel, G.I, White, S.A, Jackson, J.B. | | Deposit date: | 2004-07-19 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Active-site conformational changes associated with hydride transfer in proton-translocating transhydrogenase.
Biochemistry, 43, 2004
|
|
1U2E
 
 | | Crystal Structure of the C-C bond hydrolase MhpC | | Descriptor: | 2-hydroxy-6-ketonona-2,4-dienedioic acid hydrolase, CHLORIDE ION | | Authors: | Montgomery, M.G, Dunn, G, Mohammed, F, Robertson, T, Garcia, J.-L, Coker, A, Bugg, T.D.H, Wood, S.P. | | Deposit date: | 2004-07-19 | | Release date: | 2005-02-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the C-C Bond Hydrolase MhpC Provides Insights into its Catalytic Mechanism
J.Mol.Biol., 346, 2005
|
|
1U2F
 
 | | SOLUTION STRUCTURE OF THE FIRST RNA-BINDING DOMAIN OF HU2AF65 | | Descriptor: | PROTEIN (SPLICING FACTOR U2AF 65 KD SUBUNIT) | | Authors: | Ito, T, Muto, Y, Green, M.R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 1999-05-26 | | Release date: | 1999-08-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the first and second RNA-binding domains of human U2 small nuclear ribonucleoprotein particle auxiliary factor (U2AF(65)).
EMBO J., 18, 1999
|
|
1U2G
 
 | | transhydrogenase (dI.ADPr)2(dIII.NADPH)1 asymmetric complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, GLYCEROL, NAD(P) transhydrogenase subunit alpha part 1, ... | | Authors: | Mather, O.C, Singh, A, van Boxel, G.I, White, S.A, Jackson, J.B. | | Deposit date: | 2004-07-19 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Active-site conformational changes associated with hydride transfer in proton-translocating transhydrogenase.
Biochemistry, 43, 2004
|
|
1U2H
 
 | | X-ray Structure of the N-terminally truncated human APEP-1 | | Descriptor: | Aortic preferentially expressed protein 1 | | Authors: | Manjasetty, B.A, Scheich, C, Roske, Y, Niesen, F.H, Gotz, F, Bussow, K, Heinemann, U. | | Deposit date: | 2004-07-19 | | Release date: | 2005-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | X-ray structure of engineered human Aortic Preferentially Expressed Protein-1 (APEG-1)
Bmc Struct.Biol., 5, 2005
|
|
1U2J
 
 | | Crystal structure of the C-terminal domain from the catalase-peroxidase KatG of Escherichia coli (P21 21 21) | | Descriptor: | Peroxidase/catalase HPI | | Authors: | Carpena, X, Melik-Adamyan, W, Loewen, P.C, Fita, I. | | Deposit date: | 2004-07-19 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the C-terminal domain of the catalase-peroxidase KatG from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1U2K
 
 | | Crystal structure of the C-terminal domain from the catalase-peroxidase KatG of Escherichia coli (I41) | | Descriptor: | Peroxidase/catalase HPI | | Authors: | Carpena, X, Melik-Adamyan, W, Loewen, P.C, Fita, I. | | Deposit date: | 2004-07-19 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the C-terminal domain of the catalase-peroxidase KatG from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1U2L
 
 | | Crystal structure of the C-terminal domain from the catalase-peroxidase KatG of Escherichia coli (P1) | | Descriptor: | Peroxidase/catalase HPI | | Authors: | Carpena, X, Melik-Adamyan, W, Loewen, P.C, Fita, I. | | Deposit date: | 2004-07-19 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the C-terminal domain of the catalase-peroxidase KatG from Escherichia coli.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1U2M
 
 | | Crystal Structure of Skp | | Descriptor: | Histone-like protein HLP-1 | | Authors: | Walton, T.A, Sousa, M.C. | | Deposit date: | 2004-07-19 | | Release date: | 2004-08-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Skp, a Prefoldin-like Chaperone that Protects Soluble and Membrane Proteins from Aggregation
Mol.Cell, 15, 2004
|
|
1U2N
 
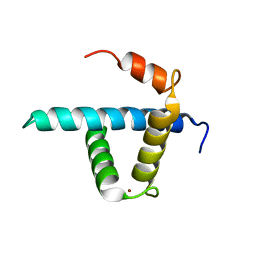 | | Structure CBP TAZ1 Domain | | Descriptor: | CREB binding protein, ZINC ION | | Authors: | De Guzman, R.N, Wojciak, J.M, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2004-07-19 | | Release date: | 2005-04-26 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | CBP/p300 TAZ1 domain forms a structured scaffold for ligand binding
Biochemistry, 44, 2005
|
|
