1BRM
 
 | | ASPARTATE BETA-SEMIALDEHYDE DEHYDROGENASE FROM ESCHERICHIA COLI | | Descriptor: | ASPARTATE-SEMIALDEHYDE DEHYDROGENASE | | Authors: | Hadfield, A.T, Kryger, G, Ouyang, J, Ringe, D, Petsko, G.A, Viola, R.E. | | Deposit date: | 1998-08-24 | | Release date: | 1999-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of aspartate-beta-semialdehyde dehydrogenase from Escherichia coli, a key enzyme in the aspartate family of amino acid biosynthesis.
J.Mol.Biol., 289, 1999
|
|
1BRN
 
 | |
1BRO
 
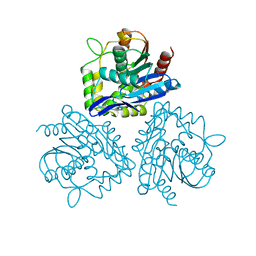 | | BROMOPEROXIDASE A2 | | Descriptor: | BROMOPEROXIDASE A2 | | Authors: | Hecht, H.J, Sobek, H, Haag, T, Pfeifer, O, Van Pee, K.H. | | Deposit date: | 1996-06-01 | | Release date: | 1996-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The metal-ion-free oxidoreductase from Streptomyces aureofaciens has an alpha/beta hydrolase fold.
Nat.Struct.Biol., 1, 1994
|
|
1BRP
 
 | |
1BRQ
 
 | |
1BRR
 
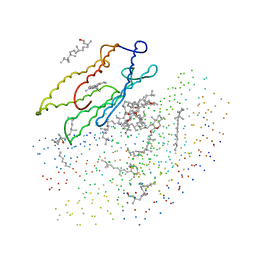 | | X-RAY STRUCTURE OF THE BACTERIORHODOPSIN TRIMER/LIPID COMPLEX | | Descriptor: | 3,7,11,15-TETRAMETHYL-HEXADECAN-1-OL, 3-O-sulfo-beta-D-galactopyranose-(1-6)-alpha-D-mannopyranose-(1-2)-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Essen, L.-O, Siegert, R, Oesterhelt, D. | | Deposit date: | 1998-07-28 | | Release date: | 1998-09-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Lipid patches in membrane protein oligomers: crystal structure of the bacteriorhodopsin-lipid complex
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1BRS
 
 | |
1BRT
 
 | | BROMOPEROXIDASE A2 MUTANT M99T | | Descriptor: | BROMOPEROXIDASE A2, CHLORIDE ION | | Authors: | Hofmann, B, Toelzer, S, Pelletier, I, Altenbuchner, J, Van Pee, K.H, Hecht, H.J. | | Deposit date: | 1998-03-30 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural investigation of the cofactor-free chloroperoxidases.
J.Mol.Biol., 279, 1998
|
|
1BRU
 
 | |
1BRV
 
 | | SOLUTION NMR STRUCTURE OF THE IMMUNODOMINANT REGION OF PROTEIN G OF BOVINE RESPIRATORY SYNCYTIAL VIRUS, 48 STRUCTURES | | Descriptor: | PROTEIN G | | Authors: | Doreleijers, J.F, Langedijk, J.P.M, Hard, K, Rullmann, J.A.C, Boelens, R, Schaaper, W.M, Van Oirschot, J.T, Kaptein, R. | | Deposit date: | 1996-03-29 | | Release date: | 1997-06-05 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the immunodominant region of protein G of bovine respiratory syncytial virus.
Biochemistry, 35, 1996
|
|
1BRW
 
 | |
1BRX
 
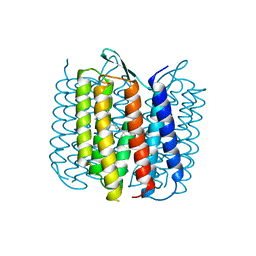 | | BACTERIORHODOPSIN/LIPID COMPLEX | | Descriptor: | BACTERIORHODOPSIN, RETINAL | | Authors: | Luecke, H, Richter, H.T, Lanyi, J. | | Deposit date: | 1998-05-28 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Proton transfer pathways in bacteriorhodopsin at 2.3 angstrom resolution.
Science, 280, 1998
|
|
1BRY
 
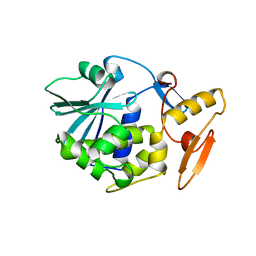 | | BRYODIN TYPE I RIP | | Descriptor: | BRYODIN I | | Authors: | Klei, H.E, Chang, C.Y. | | Deposit date: | 1997-02-14 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular, biological, and preliminary structural analysis of recombinant bryodin 1, a ribosome-inactivating protein from the plant Bryonia dioica.
Biochemistry, 36, 1997
|
|
1BRZ
 
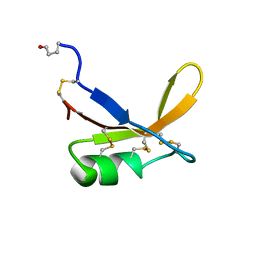 | | SOLUTION STRUCTURE OF THE SWEET PROTEIN BRAZZEIN, NMR, 43 STRUCTURES | | Descriptor: | BRAZZEIN | | Authors: | Caldwell, J.E, Abildgaard, F, Dzakula, Z, Ming, D, Hellekant, G, Markley, J.L. | | Deposit date: | 1998-03-12 | | Release date: | 1998-07-01 | | Last modified: | 2019-12-25 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the thermostable sweet-tasting protein brazzein.
Nat.Struct.Biol., 5, 1998
|
|
1BS0
 
 | | PLP-DEPENDENT ACYL-COA SYNTHASE | | Descriptor: | PROTEIN (8-AMINO-7-OXONANOATE SYNTHASE), SULFATE ION | | Authors: | Alexeev, D, Alexeeva, M, Baxter, R.L, Campopiano, D.J, Webster, S.P, Sawyer, L. | | Deposit date: | 1998-08-31 | | Release date: | 1999-08-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of 8-amino-7-oxononanoate synthase: a bacterial PLP-dependent, acyl-CoA-condensing enzyme.
J.Mol.Biol., 284, 1998
|
|
1BS1
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH DETHIOBIOTIN, ADP , INORGANIC PHOSPHATE AND MAGNESIUM | | Descriptor: | 8-AMINO-7-CARBOXYAMINO-NONANOIC ACID WITH ALUMINUM FLUORIDE, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kaeck, H, Sandmark, J, Gibson, K.J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1998-08-31 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of two quaternary complexes of dethiobiotin synthetase, enzyme-MgADP-AlF3-diaminopelargonic acid and enzyme-MgADP-dethiobiotin-phosphate; implications for catalysis.
Protein Sci., 7, 1998
|
|
1BS2
 
 | | YEAST ARGINYL-TRNA SYNTHETASE | | Descriptor: | ARGININE, PROTEIN (ARGINYL-TRNA SYNTHETASE) | | Authors: | Cavarelli, J, Delagouute, B, Eriani, G, Gangloff, J, Moras, D. | | Deposit date: | 1998-08-31 | | Release date: | 1999-08-27 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | L-arginine recognition by yeast arginyl-tRNA synthetase.
EMBO J., 17, 1998
|
|
1BS3
 
 | | P.SHERMANII SOD(FE+3) FLUORIDE | | Descriptor: | FE (III) ION, FLUORIDE ION, SUPEROXIDE DISMUTASE | | Authors: | Schmidt, M. | | Deposit date: | 1998-08-31 | | Release date: | 1999-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Manipulating the coordination mumber of the ferric iron within the cambialistic superoxide dismutase of Propionibacterium shermanii by changing the pH-value A crystallographic analysis
Eur.J.Biochem., 262, 1999
|
|
1BS4
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM (NATIVE) IN COMPLEX WITH INHIBITOR POLYETHYLENE GLYCOL | | Descriptor: | NONAETHYLENE GLYCOL, PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS5
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM | | Descriptor: | PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ZINC ION | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS6
 
 | | PEPTIDE DEFORMYLASE AS NI2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | Descriptor: | NICKEL (II) ION, PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS7
 
 | | PEPTIDE DEFORMYLASE AS NI2+ CONTAINING FORM | | Descriptor: | NICKEL (II) ION, PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of peptide deformylase and identification of the substrate binding site.
J.Biol.Chem., 273, 1998
|
|
1BS8
 
 | | PEPTIDE DEFORMYLASE AS ZN2+ CONTAINING FORM IN COMPLEX WITH TRIPEPTIDE MET-ALA-SER | | Descriptor: | PROTEIN (MET-ALA-SER), PROTEIN (PEPTIDE DEFORMYLASE), SULFATE ION, ... | | Authors: | Becker, A, Schlichting, I, Kabsch, W, Groche, D, Schultz, S, Wagner, A.F.V. | | Deposit date: | 1998-09-01 | | Release date: | 1999-08-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Iron center, substrate recognition and mechanism of peptide deformylase.
Nat.Struct.Biol., 5, 1998
|
|
1BS9
 
 | | ACETYLXYLAN ESTERASE FROM P. PURPUROGENUM REFINED AT 1.10 ANGSTROMS | | Descriptor: | ACETYL XYLAN ESTERASE, SULFATE ION | | Authors: | Ghosh, D, Erman, M, Sawicki, M.W, Lala, P, Weeks, D.R, Li, N, Pangborn, W, Thiel, D.J, Jornvall, H, Eyzaguirre, J. | | Deposit date: | 1998-09-01 | | Release date: | 1999-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Determination of a protein structure by iodination: the structure of iodinated acetylxylan esterase.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1BSA
 
 | |
