2IJD
 
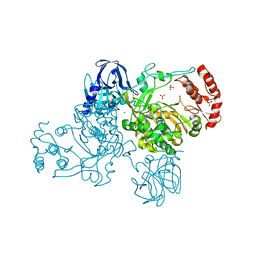 | | Crystal Structure of the Poliovirus Precursor Protein 3CD | | Descriptor: | Picornain 3C, RNA-directed RNA polymerase, SULFATE ION, ... | | Authors: | Marcotte, L.L, Gohara, D.W, Filman, D.J, Hogle, J.M. | | Deposit date: | 2006-09-29 | | Release date: | 2007-02-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of poliovirus 3CD: virally-encoded protease and precursor to the RNA-dependent RNA polymerase.
J.Virol., 81, 2007
|
|
8SP9
 
 | |
5MM2
 
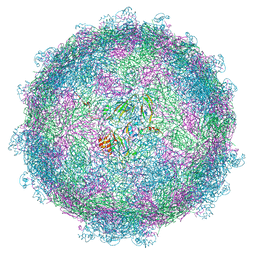 | | nora virus structure | | Descriptor: | Capsid protein VP4A, capsid protein VP4B, capsid protein VP4C | | Authors: | Laurinmaki, P, Shakeel, S, Ekstrom, J.-O, Butcher, S.J. | | Deposit date: | 2016-12-08 | | Release date: | 2017-12-20 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structure of Nora virus at 2.7 angstrom resolution and implications for receptor binding, capsid stability and taxonomy.
Sci Rep, 10, 2020
|
|
2X5I
 
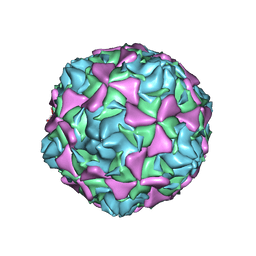 | | Crystal structure echovirus 7 | | Descriptor: | LAURIC ACID, VP1, VP2, ... | | Authors: | Plevka, P, Hafenstein, S, Zhang, Y, Bowman, V.D, Chipman, P.R, Bator, C.M, Rossmann, M.G. | | Deposit date: | 2010-02-08 | | Release date: | 2010-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Interaction of Decay-Accelerating Factor with Echovirus 7.
J.Virol., 84, 2010
|
|
6QWT
 
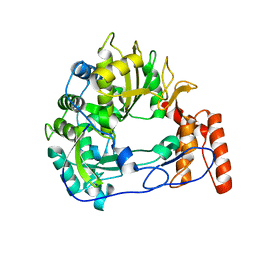 | | Sicinivirus 3Dpol RNA dependent RNA polymerase | | Descriptor: | Genome polyprotein | | Authors: | Dubankova, A, Boura, E. | | Deposit date: | 2019-03-06 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of kobuviral and siciniviral polymerases reveal conserved mechanism of picornaviral polymerase activation.
J.Struct.Biol., 208, 2019
|
|
8GOT
 
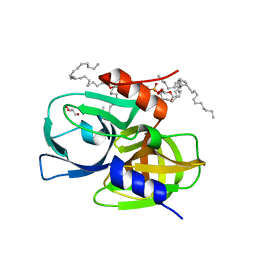 | | Crystal structure of wild-type protease 3C from Seneca Valley Virus | | Descriptor: | GLYCEROL, Peptidase C3, [(2~{S})-2-hexadecanoyloxy-3-[[(2~{R})-3-[[(2~{S})-3-[(5~{E},8~{E},11~{Z},14~{E})-icosa-5,8,11,14-tetraenoyl]oxy-2-[(9~{E},12~{Z})-octadeca-9,12-dienoyl]oxy-propoxy]-oxidanyl-phosphoryl]oxy-2-oxidanyl-propoxy]-oxidanyl-phosphoryl]oxy-propyl] icosanoate | | Authors: | Zhao, H.F, Zhang, H. | | Deposit date: | 2022-08-25 | | Release date: | 2023-05-24 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Allosteric regulation of Senecavirus A 3Cpro proteolytic activity by an endogenous phospholipid.
Plos Pathog., 19, 2023
|
|
8GPH
 
 | |
6R1I
 
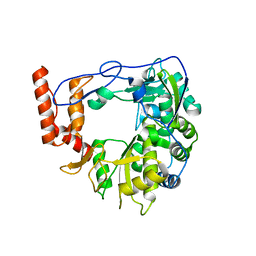 | | Structure of porcine Aichi virus polymerase | | Descriptor: | Genome polyprotein | | Authors: | Dubankova, A, Boura, E. | | Deposit date: | 2019-03-14 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.634 Å) | | Cite: | Structures of kobuviral and siciniviral polymerases reveal conserved mechanism of picornaviral polymerase activation.
J.Struct.Biol., 208, 2019
|
|
5OYP
 
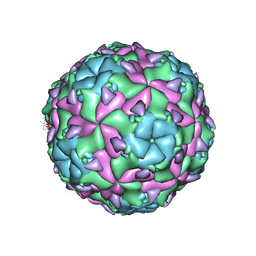 | | Sacbrood virus of honeybee | | Descriptor: | minor capsid protein MiCP, structural protein VP1, structural protein VP2, ... | | Authors: | Plevka, P, Prochazkova, M. | | Deposit date: | 2017-09-11 | | Release date: | 2018-07-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Virion structure and genome delivery mechanism of sacbrood honeybee virus.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7L8I
 
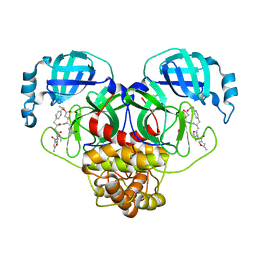 | | SARS-CoV-2 Main Protease (Mpro) in Complex with Rupintrivir (P21) | | Descriptor: | 3C-like proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Lockbaum, G.J, Henes, M, Lee, J.M, Timm, J, Nalivaika, E.A, Yilmaz, N.K, Thompson, P.R, Schiffer, C.A. | | Deposit date: | 2020-12-31 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode.
Biochemistry, 60, 2021
|
|
7L8J
 
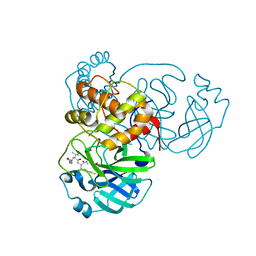 | | SARS-CoV-2 Main Protease (Mpro) in Complex with Rupintrivir (P21212) | | Descriptor: | 3C-like proteinase, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Lockbaum, G.J, Henes, M, Lee, J.M, Timm, J, Nalivaika, E.A, Yilmaz, N.K, Thompson, P.R, Schiffer, C.A. | | Deposit date: | 2020-12-31 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode.
Biochemistry, 60, 2021
|
|
7L8H
 
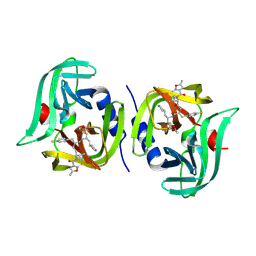 | | EV68 3C protease (3Cpro) in Complex with Rupintrivir | | Descriptor: | 3C Protease, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Lockbaum, G.J, Henes, M, Lee, J.M, Timm, J, Nalivaika, E.A, Yilmaz, N.K, Thompson, P.R, Schiffer, C.A. | | Deposit date: | 2020-12-31 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pan-3C Protease Inhibitor Rupintrivir Binds SARS-CoV-2 Main Protease in a Unique Binding Mode.
Biochemistry, 60, 2021
|
|
6ES8
 
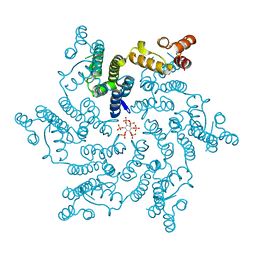 | | HIV capsid hexamer with IP6 ligand | | Descriptor: | Gag protein, INOSITOL HEXAKISPHOSPHATE | | Authors: | James, L.C. | | Deposit date: | 2017-10-19 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis.
Elife, 7, 2018
|
|
6EIT
 
 | |
3BSO
 
 | |
3BSN
 
 | |
7BZT
 
 | | Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BZN
 
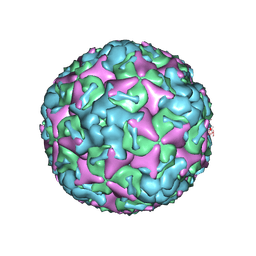 | | Cryo-EM structure of mature Coxsackievirus A10 at pH 7.4 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BZO
 
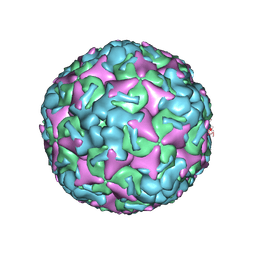 | | Cryo-EM structure of mature Coxsackievirus A10 at pH 5.5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7BZU
 
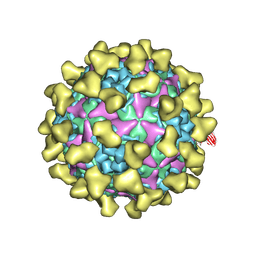 | | Cryo-EM structure of mature Coxsackievirus A10 in complex with KRM1 at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Cui, Y, Peng, R, Song, H, Tong, Z, Gao, G.F, Qi, J. | | Deposit date: | 2020-04-28 | | Release date: | 2020-07-22 | | Last modified: | 2020-08-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
1AL2
 
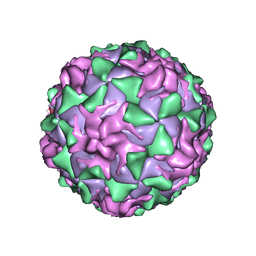 | | P1/MAHONEY POLIOVIRUS, SINGLE SITE MUTANT V1160I | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-06-09 | | Release date: | 1997-11-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1AR7
 
 | | P1/MAHONEY POLIOVIRUS, DOUBLE MUTANT P1095S + H2142Y | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1ASJ
 
 | | P1/MAHONEY POLIOVIRUS, AT CRYOGENIC TEMPERATURE | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
1AR8
 
 | | P1/MAHONEY POLIOVIRUS, MUTANT P1095S | | Descriptor: | MYRISTIC ACID, P1/MAHONEY POLIOVIRUS, SPHINGOSINE | | Authors: | Wien, M.W, Curry, S, Filman, D.J, Hogle, J.M. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural studies of poliovirus mutants that overcome receptor defects.
Nat.Struct.Biol., 4, 1997
|
|
3IYB
 
 | | Poliovirus early RNA-release intermediate | | Descriptor: | Genome polyprotein, Precursor polyprotein, VP1 core | | Authors: | Levy, H.C, Bostina, M, Filman, D.J, Hogle, J.M. | | Deposit date: | 2009-07-21 | | Release date: | 2010-03-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy.
J.Virol., 84, 2010
|
|
