5Q04
 
 | | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the allosteric inhibitor 1-(4-bromo-5-chlorothiophen-2-yl)sulfonyl-3-(5-bromo-1,3-thiazol-2-yl)urea | | Descriptor: | 4-bromo-N-[(5-bromo-1,3-thiazol-2-yl)carbamoyl]-5-chlorothiophene-2-sulfonamide, Fructose-1,6-bisphosphatase 1 | | Authors: | Ruf, A, Joseph, C, Alker, A, Banner, D, Tetaz, T, Benz, J, Kuhn, B, Rudolph, M.G, Yang, H, Shao, C, Burley, S.K. | | Deposit date: | 2017-04-18 | | Release date: | 2019-01-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Human liver fructose-1,6-bisphosphatase 1 (fructose 1,6-bisphosphate 1-phosphatase, E.C.3.1.3.11) complexed with the
allosteric inhibitor 1-(4-bromo-5-chlorothiophen-2-yl)sulfonyl-3-(5-bromo-1,3-thiazol-2-yl)urea
To be published
|
|
1UXN
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-02-27 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
6MZC
 
 | | Human TFIID BC core | | Descriptor: | Transcription initiation factor TFIID subunit 10, Transcription initiation factor TFIID subunit 12, Transcription initiation factor TFIID subunit 2, ... | | Authors: | Patel, A.B, Louder, R.K, Greber, B.J, Grunberg, S, Luo, J, Fang, J, Liu, Y, Ranish, J, Hahn, S, Nogales, E. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure of human TFIID and mechanism of TBP loading onto promoter DNA.
Science, 362, 2018
|
|
6KAD
 
 | | Cryo-EM structure of the C2S2M2L2-type PSII-LHCII supercomplex from Chlamydomonas reihardtii | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Sheng, X, Watanabe, A, Li, A.J, Kim, E, Song, C, Murata, K, Song, D.F, Minagawa, J, Liu, Z.F. | | Deposit date: | 2019-06-21 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insight into light harvesting for photosystem II in green algae.
Nat.Plants, 5, 2019
|
|
7M4Z
 
 | | A. baumannii Ribosome-Eravacycline complex: hpf-bound 70S | | Descriptor: | 16s Ribosomal RNA, 23s ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
3ZI5
 
 | | Crystal STRUCTURE OF RESTRICTION ENDONUCLEASE BFII C-TERMINAL RECOGNITION DOMAIN IN COMPLEX WITH COGNATE DNA | | Descriptor: | 5'-D(*AP*GP*CP*AP*CP*TP*GP*GP*GP*TP*CP*GP)-3', 5'-D(*CP*GP*AP*CP*CP*CP*AP*GP*TP*GP*CP*TP)-3', RESTRICTION ENDONUCLEASE | | Authors: | Golovenko, D, Manakova, E, Zakrys, L, Zaremba, M, Sasnauskas, G, Grazulis, S, Siksnys, V. | | Deposit date: | 2013-01-03 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insight Into the Specificity of the B3 DNA-Binding Domains Provided by the Co-Crystal Structure of the C-Terminal Fragment of Bfii Restriction Enzyme
Nucleic Acids Res., 42, 2014
|
|
2X6M
 
 | | Structure of a single domain camelid antibody fragment in complex with a C-terminal peptide of alpha-synuclein | | Descriptor: | ALPHA-SYNUCLEIN PEPTIDE, HEAVY CHAIN VARIABLE DOMAIN FROM DROMEDARY | | Authors: | DeGenst, E, Guilliams, T, Wellens, J, O'Day, E.M, Waudby, C.A, Meehan, S, Dumoulin, M, Hsu, S.-T.D, Cremades, N, Verschueren, K.H.G, Pardon, E, Wyns, L, Steyaert, J, Christodoulou, J, Dobson, C.M. | | Deposit date: | 2010-02-18 | | Release date: | 2010-06-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure and Properties of a Complex of Alpha-Synuclein and a Single-Domain Camelid Antibody.
J.Mol.Biol., 402, 2010
|
|
4CDQ
 
 | | Crystal structure of human Enterovirus 71 in complex with the uncoating inhibitor GPP2 | | Descriptor: | 4-((5-(2-oxo-3-(pyridin-4-yl)imidazolidin-1-yl)pentyl)oxy)benzaldehyde O-ethyl oxime, SODIUM ION, VP1, ... | | Authors: | DeColibus, L, Wang, X, Spyrou, J.A.B, Kelly, J, Ren, J, Grimes, J, Puerstinger, G, Stonehouse, N, Walter, T.S, Hu, Z, Wang, J, Li, X, Peng, W, Rowlands, D, Fry, E.E, Rao, Z, Stuart, D.I. | | Deposit date: | 2013-11-05 | | Release date: | 2014-02-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | More-Powerful Virus Inhibitors from Structure-Based Analysis of Hev71 Capsid-Binding Molecules
Nat.Struct.Mol.Biol., 21, 2014
|
|
7M4V
 
 | | A. baumannii Ribosome-Eravacycline complex: 50S | | Descriptor: | 23s ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
7M4W
 
 | | A. baumannii Ribosome-Eravacycline complex: Empty 70S | | Descriptor: | 16s Ribosomal RNA, 23s ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Morgan, C.E, Yu, E.W. | | Deposit date: | 2021-03-22 | | Release date: | 2021-05-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Cryo-EM Determination of Eravacycline-Bound Structures of the Ribosome and the Multidrug Efflux Pump AdeJ of Acinetobacter baumannii.
Mbio, 12, 2021
|
|
6H3Z
 
 | | Crystal structure of a C-terminal MIF4G domain in NOT1 | | Descriptor: | CCR4-not transcription complex subunit 1, SODIUM ION | | Authors: | Raisch, T, Sandmeir, F, Weichenrieder, O, Valkov, E, Izaurralde, E. | | Deposit date: | 2018-07-19 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and biochemical analysis of a NOT1 MIF4G-like domain of the CCR4-NOT complex.
J. Struct. Biol., 204, 2018
|
|
5EA7
 
 | | Crystal Structure of Inhibitor BMS-433771 in Complex with Prefusion RSV F Glycoprotein | | Descriptor: | 1-cyclopropyl-3-[[1-(4-oxidanylbutyl)benzimidazol-2-yl]methyl]imidazo[4,5-c]pyridin-2-one, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Fusion glycoprotein F0, ... | | Authors: | Battles, M.B, McLellan, J.S, Arnoult, E, Roymans, D, Langedijk, J.P. | | Deposit date: | 2015-10-15 | | Release date: | 2015-12-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Molecular mechanism of respiratory syncytial virus fusion inhibitors.
Nat.Chem.Biol., 12, 2016
|
|
1NWI
 
 | |
7M8I
 
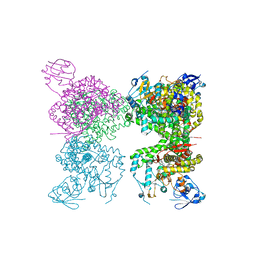 | | Human CYP11B2 and human adrenodoxin in complex with fadrozole | | Descriptor: | 4-[(5R)-5,6,7,8-tetrahydroimidazo[1,5-a]pyridin-5-yl]benzonitrile, Adrenodoxin, Cytochrome P450 11B2, ... | | Authors: | Scott, E.E, Brixius-Anderko, S. | | Deposit date: | 2021-03-29 | | Release date: | 2021-05-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structural and functional insights into aldosterone synthase interaction with its redox partner protein adrenodoxin.
J.Biol.Chem., 296, 2021
|
|
1UVH
 
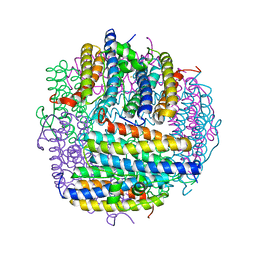 | | X-ray structure of Dps from Mycobacterium smegmatis | | Descriptor: | FE (III) ION, STARVATION-INDUCED DNA PROTECTING PROTEIN | | Authors: | Ilari, A, Ceci, P, Falvo, E, Chiancone, E. | | Deposit date: | 2004-01-20 | | Release date: | 2005-02-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Reassessment of Protein Stability, DNA Binding, and Protection of Mycobacterium Smegmatis Dps.
J.Biol.Chem., 280, 2005
|
|
3CT2
 
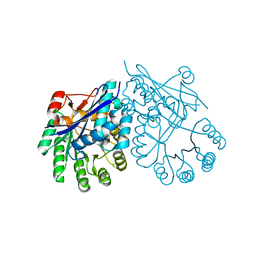 | | Crystal structure of muconate cycloisomerase from Pseudomonas fluorescens | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-04-11 | | Release date: | 2008-04-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis,cis-muconate lactonizing enzymes.
Biochemistry, 48, 2009
|
|
4NT2
 
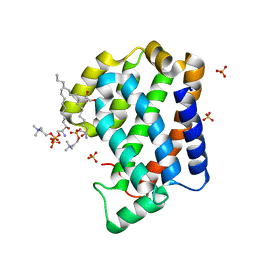 | | Crystal structure of Arabidopsis ACD11 (accelerated-cell-death 11) complexed with lyso-sphingomyelin (d18:1) at 2.4 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(R)-{[(2S,3R,4E)-2-amino-3-hydroxyoctadec-4-en-1-yl]oxy}(hydroxy)phosphoryl]oxy}-N,N,N-trimethylethanaminium, SULFATE ION, ... | | Authors: | Simanshu, D.K, Brown, R.E, Patel, D.J. | | Deposit date: | 2013-11-29 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Arabidopsis Accelerated Cell Death 11, ACD11, Is a Ceramide-1-Phosphate Transfer Protein and Intermediary Regulator of Phytoceramide Levels.
Cell Rep, 6, 2014
|
|
6MS8
 
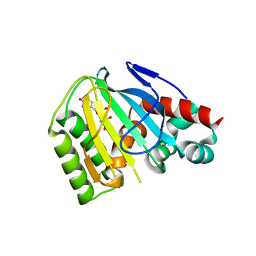 | | Crystal Structure of Chalcone Isomerase from Medicago Truncatula Complexed with (2S) Naringenin | | Descriptor: | Chalcone-flavonone isomerase family protein, NARINGENIN | | Authors: | Burke, J.R, La Clair, J.J, Philippe, R.N, Pabis, A, Jez, J.M, Cortina, G, Kaltenbach, M, Bowman, M.E, Woods, K.B, Nelson, A.T, Tawfik, D.S, Kamerlin, S.C.L, Noel, J.P. | | Deposit date: | 2018-10-16 | | Release date: | 2019-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Bifunctional Substrate Activation via an Arginine Residue Drives Catalysis in Chalcone Isomerases
Acs Catalysis, 2019
|
|
6D20
 
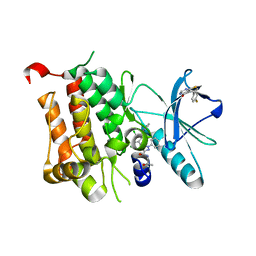 | | Crystal structure of Tyrosine-protein kinase receptor in complex with 5-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4(3H)-one and 5-{[2,4-dichloro-5-(pyridin-2-yl)benzene-1-carbonyl]amino}-N-(2-hydroxy-2-methylpropyl)-1-phenyl-1H-pyrazole-3-carboxamide Inhibitors | | Descriptor: | 5-(4-fluorophenyl)thieno[2,3-d]pyrimidin-4(3H)-one, 5-{[2,4-dichloro-5-(pyridin-2-yl)benzene-1-carbonyl]amino}-N-(2-hydroxy-2-methylpropyl)-1-phenyl-1H-pyrazole-3-carboxamide, High affinity nerve growth factor receptor | | Authors: | Greasley, S.E, Johnson, E, Kraus, M.L, Cronin, C.N. | | Deposit date: | 2018-04-12 | | Release date: | 2018-05-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of Allosteric, Potent, Subtype Selective, and Peripherally Restricted TrkA Kinase Inhibitors.
J. Med. Chem., 62, 2019
|
|
2F8P
 
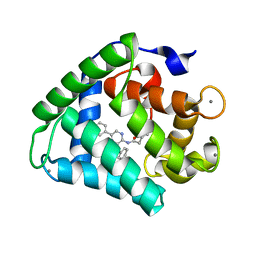 | | Crystal structure of obelin following Ca2+ triggered bioluminescence suggests neutral coelenteramide as the primary excited state | | Descriptor: | CALCIUM ION, N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE, Obelin | | Authors: | Liu, Z.J, Stepanyuk, G.A, Vysotski, E.S, Lee, J, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-12-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of obelin after Ca2+-triggered bioluminescence suggests neutral coelenteramide as the primary excited state.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
6NW1
 
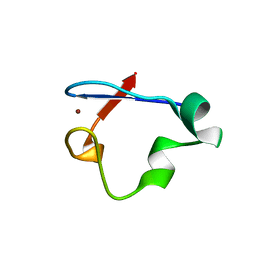 | | Crystal Structure Desulfovibrio desulfuricans Nickel-Substituted Rubredoxin V37N | | Descriptor: | NICKEL (II) ION, Rubredoxin | | Authors: | Slater, J.W, Marguet, S.C, Gray, M.E, Sotomayor, M, Shafaat, H.S. | | Deposit date: | 2019-02-05 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Power of the Secondary Sphere: Modulating Hydrogenase Activity in Nickel-Substituted Rubredoxin
Acs Catalysis, 2019
|
|
5BXV
 
 | | eIF4E complex | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4E-binding protein 1 | | Authors: | Sekiyama, N, Arthanari, H, Papdopoulos, E, Rodriguez-Mias, R.A, Wagner, G, Leger-Abraham, M. | | Deposit date: | 2015-06-09 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular mechanism of the dual activity of 4EGI-1: Dissociating eIF4G from eIF4E but stabilizing the binding of unphosphorylated 4E-BP1.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5VOV
 
 | | Structure of AMPA receptor-TARP complex | | Descriptor: | Glutamate receptor 2, Voltage-dependent calcium channel gamma-2 subunit | | Authors: | Zhao, Y, Chen, S, Wang, Y.S, Shekhar, M, Tajkhorshid, E, Gouaux, E. | | Deposit date: | 2017-05-03 | | Release date: | 2017-07-12 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Activation and Desensitization Mechanism of AMPA Receptor-TARP Complex by Cryo-EM.
Cell, 170, 2017
|
|
6NRX
 
 | | Crystal structure of DIP-eta IG1 homodimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dpr-interacting protein eta, isoform B, ... | | Authors: | Cheng, S, Park, Y.J, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-01-24 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular basis of synaptic specificity by immunoglobulin superfamily receptors in Drosophila.
Elife, 8, 2019
|
|
5C6T
 
 | | Crystal structure of HCMV glycoprotein B in complex with 1G2 Fab | | Descriptor: | 1G2 Fab heavy chain, 1G2 Fab light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chandramouli, S, Ciferri, C, Settembre, E.C, Carfi, A. | | Deposit date: | 2015-06-23 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of HCMV glycoprotein B in the postfusion conformation bound to a neutralizing human antibody.
Nat Commun, 6, 2015
|
|
