1PQQ
 
 | | NMR Structure of a Cyclic Polyamide-DNA Complex | | Descriptor: | 45-(3-AMINOPROPYL)-5,11,22,28,34-PENTAMETHYL-3,9,15,20,26,32,38,43-OCTAOXO-2,5,8,14,19,22,25,28,31,34,37,42,45,48-TETRADECAAZA-11-AZONIAHEPTACYCLO[42.2.1.1~4,7~.1~10,13~.1~21,24~.1~27,30~.1~33,36~]DOPENTACONTA-1(46),4(52),6,10(51),12,21(50),23,27(49),29,33(48),35,44(47)-DODECAENE, 5'-D(*CP*GP*CP*TP*AP*AP*CP*AP*GP*GP*C)-3', 5'-D(*GP*CP*CP*TP*GP*TP*TP*AP*GP*CP*G)-3' | | Authors: | Zhang, Q, Dwyer, T.J, Tsui, V, Case, D.A, Cho, J, Dervan, P.B, Wemmer, D.E. | | Deposit date: | 2003-06-18 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of a Cyclic Polyamide-DNA Complex.
J.Am.Chem.Soc., 126, 2004
|
|
1R7W
 
 | | NMR STRUCTURE OF THE R(GGAGGACAUCCCUCACGGGUGACCGUGGUCCUCC), DOMAIN IV STEM-LOOP B OF ENTEROVIRAL IRES WITH AUCCCU BULGE | | Descriptor: | 34-MER | | Authors: | Du, Z, Ulyanov, N.B, Yu, J, James, T.L. | | Deposit date: | 2003-10-22 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Loop B RNAs from the Stem-Loop IV Domain of the Enterovirus Internal Ribosome Entry Site: A Single C to U Substitution Drastically Changes the Shape and Flexibility of RNA(,).
Biochemistry, 43, 2004
|
|
1ZK6
 
 | | NMR solution structure of B. subtilis PrsA PPIase | | Descriptor: | Foldase protein prsA | | Authors: | Tossavainen, H, Permi, P, Purhonen, S.L, Sarvas, M, Kilpelainen, I, Seppala, R. | | Deposit date: | 2005-05-02 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and characterization of substrate binding site of the PPIase domain of PrsA protein from Bacillus subtilis
Febs Lett., 580, 2006
|
|
1YLG
 
 | | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor | | Descriptor: | apolipoprotein B mRNA | | Authors: | Maris, C, Masse, J, Allain, F.H, Chester, A, Navaratnam, N. | | Deposit date: | 2005-01-19 | | Release date: | 2005-02-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the apoB mRNA stem-loop and its interaction with the C to U editing APOBEC1 complementary factor.
Rna, 11, 2005
|
|
1RGJ
 
 | | NMR STRUCTURE OF THE COMPLEX BETWEEN ALPHA-BUNGAROTOXIN AND MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR WITH ENHANCED ACTIVITY | | Descriptor: | MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR, long neurotoxin 1 | | Authors: | Bernini, A, Spiga, O, Ciutti, A, Scarselli, M, Bracci, L, Lozzi, L, Lelli, B, Neri, P, Niccolai, N. | | Deposit date: | 2003-11-12 | | Release date: | 2003-11-25 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR and MD studies on the interaction between ligand peptides and alpha-bungarotoxin.
J.Mol.Biol., 339, 2004
|
|
1XWH
 
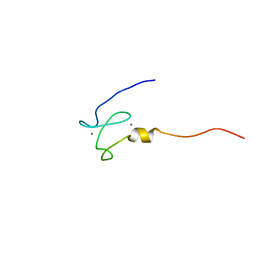 | | NMR structure of the first phd finger of autoimmune regulator protein (AIRE1): insights into apeced | | Descriptor: | Autoimmune regulator, ZINC ION | | Authors: | Bottomley, M.J, Stier, G, Krasotkina, J, Legube, G, Simon, B, Akhtar, A, Sattler, M, Musco, G. | | Deposit date: | 2004-11-01 | | Release date: | 2005-01-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the first PHD finger of autoimmune regulator protein (AIRE1). Insights into autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED) disease
J.Biol.Chem., 280, 2005
|
|
1S05
 
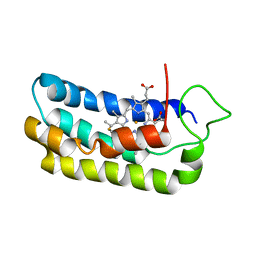 | | NMR-validated structural model for oxidized R.palustris cytochrome c556 | | Descriptor: | Cytochrome c-556, HEME C | | Authors: | Bertini, I, Faraone-Mennella, J, Gray, H.B, Luchinat, C, Parigi, G, Winkler, J.R. | | Deposit date: | 2003-12-30 | | Release date: | 2004-01-20 | | Last modified: | 2021-03-03 | | Method: | SOLUTION NMR | | Cite: | NMR-validated structural model for oxidized Rhodopseudomonas palustris cytochrome c(556).
J.Biol.Inorg.Chem., 9, 2004
|
|
1JBD
 
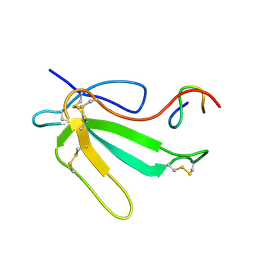 | | NMR Structure of the Complex Between alpha-bungarotoxin and a Mimotope of the Nicotinic Acetylcholine Receptor | | Descriptor: | LONG NEUROTOXIN 1, MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR | | Authors: | Scarselli, M, Spiga, O, Ciutti, A, Bracci, L, Lelli, B, Lozzi, L, Calamandrei, D, Bernini, A, Di Maro, D, Klein, S, Niccolai, N. | | Deposit date: | 2001-06-04 | | Release date: | 2001-06-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of alpha-bungarotoxin free and bound to a mimotope of the nicotinic acetylcholine receptor.
Biochemistry, 41, 2002
|
|
1PC0
 
 | |
1PYV
 
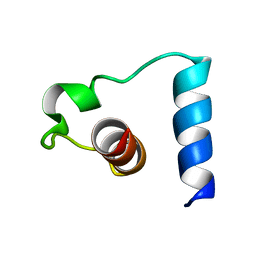 | | NMR solution structure of the mitochondrial F1b presequence peptide from Nicotiana plumbaginifolia | | Descriptor: | ATP synthase beta chain, mitochondrial precursor | | Authors: | Moberg, P, Nilsson, S, Stahl, A, Eriksson, A.C, Glaser, E, Maler, L. | | Deposit date: | 2003-07-09 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of the mitochondrial F1beta presequence from Nicotiana plumbaginifolia
J.Mol.Biol., 336, 2004
|
|
1QS3
 
 | | NMR SOLUTION CONFORMATION OF AN ANTITOXIC ANALOG OF ALPHA-CONOTOXIN GI | | Descriptor: | DES-GLU1-[CYS3ALA]-DES-CYS13-ALPHA CONOTOXIN GI | | Authors: | Mok, K.H, Han, K.-H. | | Deposit date: | 1999-06-25 | | Release date: | 1999-10-06 | | Last modified: | 2014-03-12 | | Method: | SOLUTION NMR | | Cite: | NMR solution conformation of an antitoxic analogue of alpha-conotoxin GI: identification of a common nicotinic acetylcholine receptor alpha 1-subunit binding surface for small ligands and alpha-conotoxins.
Biochemistry, 38, 1999
|
|
1OKF
 
 | | NMR structure of an alpha-L-LNA:RNA hybrid | | Descriptor: | 5'-D(*CP*ATLP*GP*AP*ATLP*AP*ATLP*GP*CP)-3', 5'-R(*GP*CP*AP*UP*AP*UP*CP*AP*GP)-3' | | Authors: | Nielsen, J.T, Stein, P.C, Petersen, M. | | Deposit date: | 2003-07-23 | | Release date: | 2003-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of an Alpha-L-Lna:RNA Hybrid: Structural Implications for Rnase H Recognition
Nucleic Acids Res., 31, 2003
|
|
1U6F
 
 | | NMR solution structure of TcUBP1, a single RBD-unit from Trypanosoma cruzi | | Descriptor: | RNA-binding protein UBP1 | | Authors: | Volpon, L, D'orso, I, Frasch, A, Gehring, K. | | Deposit date: | 2004-07-29 | | Release date: | 2005-01-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structural Study of TcUBP1, a Single RRM Domain Protein from Trypanosoma cruzi: Contribution of a beta Hairpin to RNA Binding
Biochemistry, 44, 2005
|
|
1R7Z
 
 | | NMR STRUCTURE OF THE R(GGAGGACAUUCCUCACGGGUGACCGUGGUCCUCC), DOMAIN IV STEM-LOOP B OF ENTEROVIRAL IRES WITH AUUCCU BULGE | | Descriptor: | 34-MER | | Authors: | Du, Z, Ulyanov, N.B, Yu, J, James, T.L. | | Deposit date: | 2003-10-22 | | Release date: | 2004-05-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of Loop B RNAs from the Stem-Loop IV Domain of the Enterovirus Internal Ribosome Entry Site: A Single C to U Substitution Drastically Changes the Shape and Flexibility of RNA(,).
Biochemistry, 43, 2004
|
|
1L3G
 
 | | NMR Structure of the DNA-binding Domain of Cell Cycle Protein, Mbp1(2-124) from Saccharomyces cerevisiae | | Descriptor: | TRANSCRIPTION FACTOR Mbp1 | | Authors: | Nair, M, McIntosh, P.B, Frenkiel, T.A, Kelly, G, Taylor, I.A, Smerdon, S.J, Lane, A.N. | | Deposit date: | 2002-02-27 | | Release date: | 2003-02-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the DNA-Binding Domain of the Cell Cycle Protein Mbp1 from Saccharomyces cerevisiae
Biochemistry, 42, 2003
|
|
1I5J
 
 | |
1JWC
 
 | |
1JFJ
 
 | | NMR SOLUTION STRUCTURE OF AN EF-HAND CALCIUM BINDING PROTEIN FROM ENTAMOEBA HISTOLYTICA | | Descriptor: | CALCIUM-BINDING PROTEIN | | Authors: | Atreya, H.S, Sahu, S.C, Bhattacharya, A, Chary, K.V.R, Govil, G. | | Deposit date: | 2001-06-20 | | Release date: | 2001-12-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR derived solution structure of an EF-hand calcium-binding protein from Entamoeba Histolytica.
Biochemistry, 40, 2001
|
|
1T50
 
 | | NMR SOLUTION STRUCTURE OF APLYSIA ATTRACTIN | | Descriptor: | Attractin | | Authors: | Ravindranath, G, Xu, Y, Schein, C.H, Rajaratnam, K, Painter, S.D, Nagle, G.T, Braun, W. | | Deposit date: | 2004-04-30 | | Release date: | 2004-05-11 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of Attractin, a Water-Borne Pheromone from the Mollusk Aplysia Attractin
Biochemistry, 42, 2003
|
|
1X30
 
 | |
1X2S
 
 | |
1X2Z
 
 | |
1X2O
 
 | |
1Z2K
 
 | | NMR structure of the D1 domain of the Natural Killer Cell Receptor, 2B4 | | Descriptor: | Natural killer cell receptor 2B4 | | Authors: | Ames, J.B, Vyas, V, Lusin, J.D, Mariuzza, R. | | Deposit date: | 2005-03-08 | | Release date: | 2005-05-03 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Natural Killer Cell Receptor 2B4:
Implications for Ligand Recognition
Biochemistry, 44, 2005
|
|
1YGM
 
 | | NMR structure of Mistic | | Descriptor: | hypothetical protein BSU31320 | | Authors: | Roosild, T.P, Greenwald, J, Vega, M, Castronovo, S, Riek, R, Choe, S. | | Deposit date: | 2005-01-05 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of Mistic, a membrane-integrating protein for membrane protein expression.
Science, 307, 2005
|
|
