3ETT
 
 | | Crystal structure of a bacterial arylsulfate sulfotransferase catalytic intermediate with 4-nitrophenol bound in the active site | | Descriptor: | Arylsulfate sulfotransferase, P-NITROPHENOL, SULFATE ION | | Authors: | Malojcic, G, Owen, R.L, Grimshaw, J.P, Glockshuber, R. | | Deposit date: | 2008-10-08 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural and biochemical basis for PAPS-independent sulfuryl transfer by aryl sulfotransferase from uropathogenic Escherichia coli.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3ETS
 
 | | Crystal structure of a bacterial arylsulfate sulfotransferase catalytic intermediate with 4-methylumbelliferone bound in the active site | | Descriptor: | 7-hydroxy-4-methyl-2H-chromen-2-one, Arylsulfate sulfotransferase, SULFATE ION | | Authors: | Malojcic, G, Owen, R.L, Grimshaw, J.P, Glockshuber, R. | | Deposit date: | 2008-10-08 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural and biochemical basis for PAPS-independent sulfuryl transfer by aryl sulfotransferase from uropathogenic Escherichia coli.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3Q40
 
 | |
3ELQ
 
 | | Crystal structure of a bacterial arylsulfate sulfotransferase | | Descriptor: | Arylsulfate sulfotransferase, CHLORIDE ION, SULFATE ION | | Authors: | Malojcic, G, Owen, R.L, Grimshaw, J.P, Glockshuber, R. | | Deposit date: | 2008-09-23 | | Release date: | 2008-11-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and biochemical basis for PAPS-independent sulfuryl transfer by aryl sulfotransferase from uropathogenic Escherichia coli.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3Q5G
 
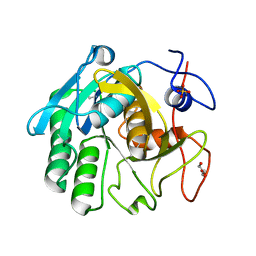 | |
2V0B
 
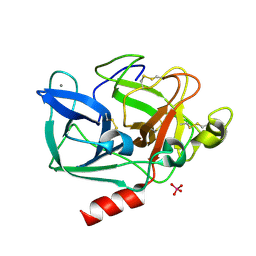 | |
2N2D
 
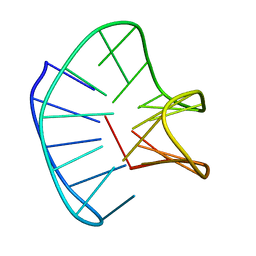 | |
2X0A
 
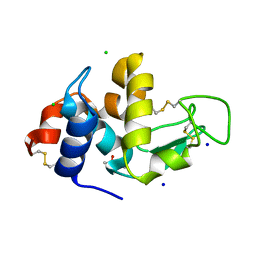 | |
2V8B
 
 | |
3L1K
 
 | |
2Q9X
 
 | | Crystal structure of highly stable mutant Q40P/S47I/H93G of human fibroblast growth factor-1 | | Descriptor: | GLYCEROL, Heparin-binding growth factor 1 | | Authors: | Szlachcic, A, Zakrzewska, M, Krowarsch, D, Os, V, Helland, R, Otlewski, J. | | Deposit date: | 2007-06-14 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of highly stable mutant Q40P/S47I/H93G of human fibroblast growth factor-1
To be Published
|
|
1W6Z
 
 | | High Energy Tetragonal Lysozyme X-ray Structure | | Descriptor: | CHLORIDE ION, HOLMIUM (III) ATOM, LYSOZYME C | | Authors: | Jakoncic, J, Aslantas, M, Honkimaki, V, Di Michiel, M, Stojanoff, V. | | Deposit date: | 2004-08-25 | | Release date: | 2004-11-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Anomalous Diffraction at Ultra-High Energy for Protein Crystallography.
J.Appl.Crystallogr., 39, 2006
|
|
1W91
 
 | |
2IWS
 
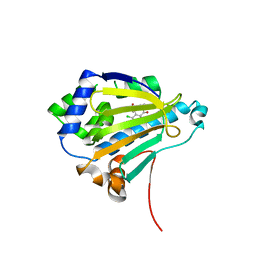 | | Radicicol analogues bound to the ATP site of HSP90 | | Descriptor: | (5Z)-12-CHLORO-13,15-DIHYDROXY-4,7,8,9-TETRAHYDRO-2-BENZOXACYCLOTRIDECINE-1,10(3H,11H)-DIONE, ATP-DEPENDENT MOLECULAR CHAPERONE HSP82 | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2006-07-04 | | Release date: | 2006-11-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibition of Hsp90 with Synthetic Macrolactones: Synthesis and Structural and Biological Evaluation of Ring and Conformational Analogs of Radicicol.
Chem.Biol., 13, 2006
|
|
2GUY
 
 | |
2JG3
 
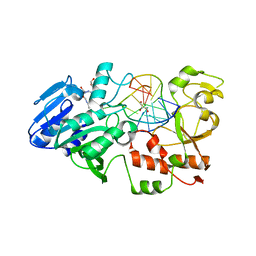 | | MtaqI with BAZ | | Descriptor: | 5'-D(*GP*AP*CP*AP*TP*CP*GP*6MAP*AP*CP)-3', 5'-D(*GP*TP*TP*CP*GP*AP*TP*GP*TP*CP)-3', 5'-DEOXY-5'-(ETHYLAMINO)-8-{[4-({5-[(3AS,4S,6AR)-2-OXOHEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL]PENTANOYL}AMINO)BUTYL]AMINO}ADENOSINE, ... | | Authors: | Pljevaljcic, G, Scheidig, A.J, Weinhold, E. | | Deposit date: | 2007-02-07 | | Release date: | 2007-08-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Quantitative Labeling of Long Plasmid DNA with Nanometer Precision.
Chembiochem, 8, 2007
|
|
2JB8
 
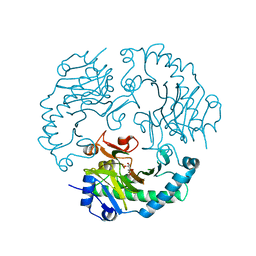 | | Deacetoxycephalosporin C synthase complexed with 5-hydroxy-4-keto valeric acid | | Descriptor: | 4,5-DIOXOPENTANOIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHETASE, FE (III) ION | | Authors: | Cicero, G, Carbonera, C, Valegard, K, Hajdu, J, Andersson, I, Ranghino, G. | | Deposit date: | 2006-12-05 | | Release date: | 2008-02-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Study of the Oxidative Half-Reaction Catalysed by a Non-Heme Ferrous Catalytic Centre by Means of Structural and Computational Methodologies
Int.J.Quantum Chem., 107, 2007
|
|
2LLU
 
 | | Post-translational S-nitrosylation is an endogenous factor fine-tuning human S100A1 protein properties | | Descriptor: | Protein S100-A1 | | Authors: | Lenarcic Zivkovic, M, Zareba-Koziol, M, Zhukova, L, Poznanski, J, Zhukov, I, Wyslouch-Cieszynska, A. | | Deposit date: | 2011-11-17 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Post-translational S-Nitrosylation Is an Endogenous Factor Fine Tuning the Properties of Human S100A1 Protein.
J.Biol.Chem., 287, 2012
|
|
2JQG
 
 | | Leader Protease | | Descriptor: | Genome polyprotein | | Authors: | Cencic, R, Mayer, C, Juliano, M.A, Juliano, L, Konrat, R, Kontaxis, G, Skern, T. | | Deposit date: | 2007-06-01 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Investigating the Substrate Specificity and Oligomerisation of the Leader Protease of Foot and Mouth Disease Virus using NMR
J.Mol.Biol., 373, 2007
|
|
2JQF
 
 | | Full Length Leader Protease of Foot and Mouth Disease Virus C51A Mutant | | Descriptor: | Genome polyprotein | | Authors: | Cencic, R, Mayer, C, Juliano, M.A, Juliano, L, Konrat, R, Kontaxis, G, Skern, T. | | Deposit date: | 2007-06-01 | | Release date: | 2007-07-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Investigating the Substrate Specificity and Oligomerisation of the Leader Protease of Foot and Mouth Disease Virus using NMR
J.Mol.Biol., 373, 2007
|
|
2LLT
 
 | | Post-translational S-nitrosylation is an endogenous factor fine-tuning human S100A1 protein properties | | Descriptor: | Protein S100-A1 | | Authors: | Lenarcic Zivkovic, M, Zareba-Koziol, M, Zhukova, L, Poznanski, J, Zhukov, I, Wyslouch-Cieszynska, A. | | Deposit date: | 2011-11-17 | | Release date: | 2012-09-26 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Post-translational S-Nitrosylation Is an Endogenous Factor Fine Tuning the Properties of Human S100A1 Protein.
J.Biol.Chem., 287, 2012
|
|
6KZG
 
 | |
1EB3
 
 | | YEAST 5-AMINOLAEVULINIC ACID DEHYDRATASE 4,7-DIOXOSEBACIC ACID COMPLEX | | Descriptor: | 4,7-DIOXOSEBACIC ACID, 5-AMINOLAEVULINIC ACID DEHYDRATASE, ZINC ION | | Authors: | Erskine, P.T, Coates, L, Newbold, R, Brindley, A.A, Stauffer, F, Wood, S.P, Warren, M.J, Cooper, J.B, Shoolingin-Jordan, P.M, Neier, R. | | Deposit date: | 2001-07-18 | | Release date: | 2001-08-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The X-Ray Structure of Yeast 5-Aminolaevulinic Acid Dehydratase Complexed with Two Diacid Inhibitors
FEBS Lett., 503, 2001
|
|
1O8W
 
 | | Radiation-reduced Tryparedoxin-I | | Descriptor: | TRYPAREDOXIN | | Authors: | Alphey, M.S, Bond, C.S, Hunter, W.N. | | Deposit date: | 2002-12-09 | | Release date: | 2003-08-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tryparedoxins from Crithidia Fasciculata and Trypanosoma Brucei: Photoreduction of the Redox Disulfide Using Synchrotron Radiation and Evidence for a Conformational Switch Implicated in Function
J.Biol.Chem., 278, 2003
|
|
5FZW
 
 | |
