6V7S
 
 | | Crystal structure of K37-acetylated SUMO1 in complex with phosphorylated PIAS-SIM2 | | Descriptor: | Protein PIAS, Small ubiquitin-related modifier 1 | | Authors: | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-12-09 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Characterization of a C-Terminal SUMO-Interacting Motif Present in Select PIAS-Family Proteins.
Structure, 28, 2020
|
|
6V7R
 
 | | Crystal structure of K37-acetylated SUMO1 in complex with PIAS-SIM2 | | Descriptor: | Protein PIAS, Small ubiquitin-related modifier 1 | | Authors: | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-12-09 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.549 Å) | | Cite: | Characterization of a C-Terminal SUMO-Interacting Motif Present in Select PIAS-Family Proteins.
Structure, 28, 2020
|
|
6V7P
 
 | | Crystal structure of SUMO1 in complex with PIAS-SIM2 | | Descriptor: | Protein PIAS, Small ubiquitin-related modifier 1 | | Authors: | Lussier-Price, M, Wahba, H.M, Mascle, X.H, Cappadocia, L, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2019-12-09 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Characterization of a C-Terminal SUMO-Interacting Motif Present in Select PIAS-Family Proteins.
Structure, 28, 2020
|
|
3MS6
 
 | | Crystal structure of Hepatitis B X-Interacting Protein (HBXIP) | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Hepatitis B virus X-interacting protein, ... | | Authors: | Garcia-Saez, I, Skoufias, D. | | Deposit date: | 2010-04-29 | | Release date: | 2010-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.085 Å) | | Cite: | Structural Characterization of HBXIP: The Protein That Interacts with the Anti-Apoptotic Protein Survivin and the Oncogenic Viral Protein HBx.
J.Mol.Biol., 405, 2011
|
|
3NJO
 
 | | X-ray crystal structure of the Pyr1-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYR1, CHLORIDE ION, ... | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Peterson, F.C, Volkman, B.F, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
6CZN
 
 | | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with Z2OHTPE and a glucocorticoid receptor-interacting protein 1 NR box II peptide | | Descriptor: | 4,4'-[(1R,2R)-1-phenylbutane-1,2-diyl]diphenol, Estrogen receptor, GRIP Peptide | | Authors: | Fanning, S.W, Han, R, Maximov, P, Jordan, V.C, Greene, G.L. | | Deposit date: | 2018-04-09 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Estrogen Receptor Alpha Ligand Binding Domain Y537S Mutant in Complex with Z2OHTPE and a glucocorticoid receptor-interacting protein 1 NR box II peptide
To Be Published
|
|
4GDN
 
 | | Structure of FmtA-like protein | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Protein flp | | Authors: | Cougnoux, A, Gibold, L, Delmas, J, Robin, F, Dalmasso, G, Bonnet, R. | | Deposit date: | 2012-08-01 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Analysis of Structure-Function Relationships in the Colibactin-Maturating Enzyme ClbP.
J.Mol.Biol., 424, 2012
|
|
8HML
 
 | |
3Q47
 
 | |
3Q49
 
 | |
8HIH
 
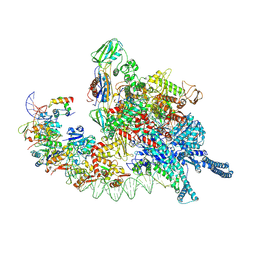 | | Cryo-EM structure of Mycobacterium tuberculosis transcription initiation complex with transcription factor GlnR | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Lin, W, Shi, J, Xu, J.C. | | Deposit date: | 2022-11-20 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural insights into the transcription activation mechanism of the global regulator GlnR from actinobacteria.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6BO0
 
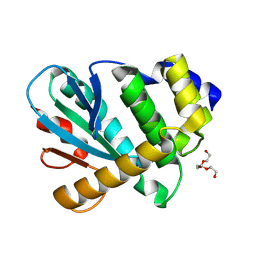 | | MdbA protein, a thiol-disulfide oxidoreductase from Corynebacterium matruchotii | | Descriptor: | MdbA protein, TETRAETHYLENE GLYCOL | | Authors: | Osipiuk, J, Luong, T.Y, Trigar, R, Ton-That, H, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-17 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Basis of a Thiol-Disulfide Oxidoreductase in the Hedgehog-Forming Actinobacterium Corynebacterium matruchotii.
J. Bacteriol., 200, 2018
|
|
6WI4
 
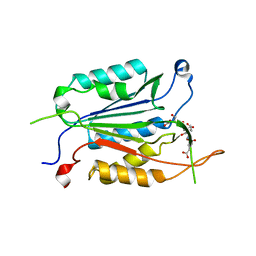 | | Caspases from Scleractinian Coral | | Descriptor: | ACE-DEVD inhibitor, Caspase-3 | | Authors: | Clark, A.C, Swartz, P.D. | | Deposit date: | 2020-04-08 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Caspases from scleractinian coral show unique regulatory features.
J.Biol.Chem., 295, 2020
|
|
4X8K
 
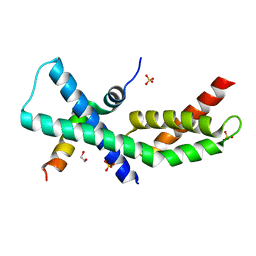 | | Mycobacterium tuberculosis RbpA-SID in complex with SigmaA domain 2 | | Descriptor: | 1,2-ETHANEDIOL, RNA polymerase sigma factor SigA, RNA polymerase-binding protein RbpA, ... | | Authors: | Hubin, E.A, Flack, J.E, Tabib-Salazar, A, Paget, M.S, Darst, S.A, Campbell, E.A. | | Deposit date: | 2014-12-10 | | Release date: | 2015-06-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structural, functional, and genetic analyses of the actinobacterial transcription factor RbpA.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3NJ1
 
 | | X-ray crystal structure of the PYL2(V114I)-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, GLYCEROL, ... | | Authors: | Peterson, F.C, Burgie, E.S, Bingman, C.A, Volkman, B.F, Phillips Jr, G.N, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NJ0
 
 | | X-ray crystal structure of the PYL2-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peterson, F.C, Burgie, E.S, Bingman, C.A, Volkman, B.F, Phillips Jr, G.N, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3OJI
 
 | | X-ray crystal structure of the Py13 -pyrabactin complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL3, SULFATE ION | | Authors: | Zhang, X, Zhang, Q, Wang, G, Chen, Z. | | Deposit date: | 2010-08-23 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complex Structures of the Abscisic Acid Receptor PYL3/RCAR13 Reveal a Unique Regulatory Mechanism
Structure, 20, 2012
|
|
8EDU
 
 | | Mycobacteriophage Muddy capsid | | Descriptor: | Capsid | | Authors: | Freeman, K.G, White, S.J, Huet, A, Conway, J.F. | | Deposit date: | 2022-09-06 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8ECO
 
 | | Microbacterium phage Oxtober96 | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8EC2
 
 | | Mycobacterium phage Adephagia | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8ECJ
 
 | | Mycobacterium phage Cain | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8ECN
 
 | | Mycobacterium phage Ogopogo | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8EB4
 
 | | Gordonia phage Ziko | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-08-30 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8ECI
 
 | | Arthrobacter phage Bridgette | | Descriptor: | Decoration protein, Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-02 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
8EC8
 
 | | Mycobacterium phage Bobi | | Descriptor: | Major capsid protein | | Authors: | Podgorski, J.M, White, S.J. | | Deposit date: | 2022-09-01 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A structural dendrogram of the actinobacteriophage major capsid proteins provides important structural insights into the evolution of capsid stability.
Structure, 31, 2023
|
|
