7OG7
 
 | | Crystal structure of the copper chaperone NosL from Shewanella denitrificans | | Descriptor: | ACETONITRILE, COPPER (I) ION, NosL, ... | | Authors: | Prasser, B, Schoener, L, Zhang, L, Einsle, O. | | Deposit date: | 2021-05-06 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Copper Chaperone NosL Forms a Heterometal Site for Cu Delivery to Nitrous Oxide Reductase.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6QHJ
 
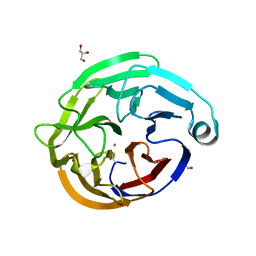 | | High-resolution crystal structure of calcium- and sodium-bound mouse Olfactomedin-1 beta-propeller domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Pronker, M.F, van den Hoek, H.G, Janssen, B.J.C. | | Deposit date: | 2019-01-16 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Design and structural characterisation of olfactomedin-1 variants as tools for functional studies.
BMC Mol Cell Biol, 20, 2019
|
|
6N7C
 
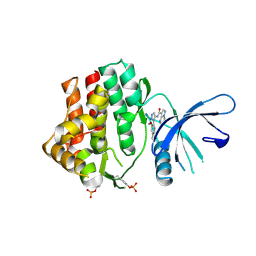 | | Structure of the human JAK1 kinase domain with compound 56 | | Descriptor: | GLYCEROL, N-[5-(3-methoxynaphthalen-2-yl)-1H-pyrazol-4-yl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6VMH
 
 | |
6QHN
 
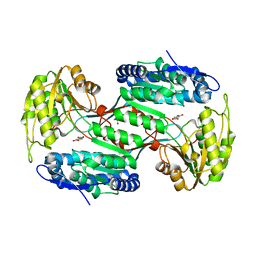 | | Metagenome-derived salicylaldehyde dehydrogenase from alpine soil in complex with protocatechuic acid | | Descriptor: | 3,4-DIHYDROXYBENZOIC ACID, GLYCEROL, SALICYLALDEHYDE DEHYDROGENASE | | Authors: | Hakansson, M, Svensson, L.A, Shamsudeen, D.U, Allen, C.C.R. | | Deposit date: | 2019-01-17 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expression, purification and crystallization of a novel metagenome-derived salicylaldehyde dehydrogenase from Alpine soil.
Acta Crystallogr.,Sect.F, 78, 2022
|
|
8BXL
 
 | | Patulin Synthase from Penicillium expansum | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Tjallinks, G, Boverio, A, Rozeboom, H.J, Fraaije, M.W. | | Deposit date: | 2022-12-09 | | Release date: | 2023-09-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure elucidation and characterization of patulin synthase, insights into the formation of a fungal mycotoxin.
Febs J., 290, 2023
|
|
7TQ4
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 6c | | Descriptor: | 3C-like proteinase, N~2~-({[(1R,2R)-2-(3-chlorophenyl)cyclopropyl]methoxy}carbonyl)-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
6BN9
 
 | | Crystal structure of DDB1-CRBN-BRD4(BD1) complex bound to dBET70 PROTAC | | Descriptor: | Bromodomain-containing protein 4, DNA damage-binding protein 1,DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Nowak, R.P, DeAngelo, S.L, Buckley, D, Bradner, J.E, Fischer, E.S. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (4.382 Å) | | Cite: | Plasticity in binding confers selectivity in ligand-induced protein degradation.
Nat. Chem. Biol., 14, 2018
|
|
6VPE
 
 | |
6QIB
 
 | |
6C3B
 
 | | O2-, PLP-Dependent L-Arginine Hydroxylase RohP Holoenzyme | | Descriptor: | 1,2-ETHANEDIOL, TRIETHYLENE GLYCOL, Uncharacterized protein | | Authors: | Hedges, J.B, Ryan, K.S. | | Deposit date: | 2018-01-09 | | Release date: | 2018-03-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Snapshots of the Catalytic Cycle of an O2, Pyridoxal Phosphate-Dependent Hydroxylase.
ACS Chem. Biol., 13, 2018
|
|
7TQ2
 
 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 1c | | Descriptor: | 3C-like proteinase, N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-({[(1R,2R)-2-phenylcyclopropyl]methoxy}carbonyl)-L-leucinamide | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
7OIL
 
 | | mPI3Kd in complex with compound 58 | | Descriptor: | 2-[(1S)-1-cyclopropylethyl]-5-[4-methyl-2-[[6-(2-oxidanylidenepyrrolidin-1-yl)pyridin-2-yl]amino]-1,3-thiazol-5-yl]-7-methylsulfonyl-3H-isoindol-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform, SODIUM ION | | Authors: | Petersen, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery of AZD8154, a Dual PI3K gamma delta Inhibitor for the Treatment of Asthma.
J.Med.Chem., 64, 2021
|
|
6N8Q
 
 | |
8C46
 
 | |
7OI4
 
 | | mPI3Kd in complex with compound 12 | | Descriptor: | N-[5-[2-[(1S)-1-cyclopropylethyl]-7-[[4-[(dimethylamino)methyl]phenyl]sulfamoyl]-1-oxidanylidene-3H-isoindol-5-yl]-4-methyl-1,3-thiazol-2-yl]ethanamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Petersen, J. | | Deposit date: | 2021-05-11 | | Release date: | 2021-07-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of AZD8154, a Dual PI3K gamma delta Inhibitor for the Treatment of Asthma.
J.Med.Chem., 64, 2021
|
|
8CHK
 
 | | Human FKBP12 in complex with (1S,5S,6R)-10-((S)-(3,5-dichlorophenyl)sulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[[3,5-bis(chloranyl)phenyl]sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Peptidyl-prolyl cis-trans isomerase FKBP1A | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
5A6M
 
 | | Determining the specificities of the catalytic site from the very high resolution structure of the thermostable glucuronoxylan endo-Beta-1, 4-xylanase, CtXyn30A, from Clostridium thermocellum with a xylotetraose bound | | Descriptor: | CARBOHYDRATE BINDING FAMILY 6, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Freire, F, Verma, A.K, Bule, P, Goyal, A, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2015-06-30 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Conservation in the Mechanism of Glucuronoxylan Hydrolysis Revealed by the Structure of Glucuronoxylan Xylano-Hydrolase (Ctxyn30A) from Clostridium Thermocellum
Acta Crystallogr.,Sect.D, 72, 2016
|
|
7WMZ
 
 | |
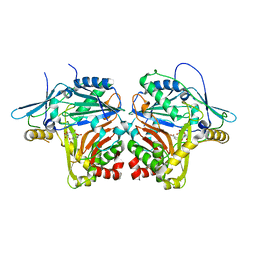
6WAO
 
 | |
7TQ3
 
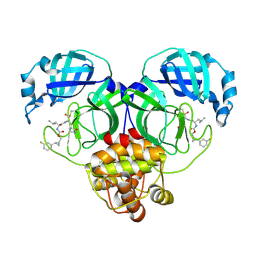 | | Structure of SARS-CoV-2 3CL protease in complex with the cyclopropane based inhibitor 5c | | Descriptor: | 3C-like proteinase, N~2~-({[(1R,2R)-2-(3-fluorophenyl)cyclopropyl]methoxy}carbonyl)-N-{(2S)-1-oxo-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Battaile, K.P, Nguyen, H.N, Chamandi, S.D, Picard, H.R, Madden, T.K, Thruman, H.A, Kim, Y, Groutas, W.C, Chang, K.O. | | Deposit date: | 2022-01-26 | | Release date: | 2022-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Broad-Spectrum Cyclopropane-Based Inhibitors of Coronavirus 3C-like Proteases: Biochemical, Structural, and Virological Studies.
Acs Pharmacol Transl Sci, 6, 2023
|
|
8CHL
 
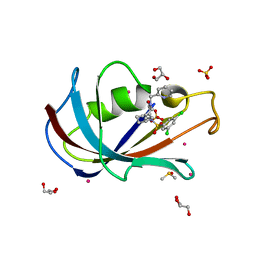 | | Human FKBP12 in complex with (1S,5S,6R)-9-((3,5-dichlorophenyl)sulfonyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,9-diazabicyclo[4.2.1]nonan-2-one | | Descriptor: | (1~{S},5~{S},6~{R})-10-[3,5-bis(chloranyl)phenyl]sulfonyl-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, CADMIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
8CHQ
 
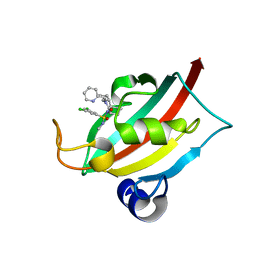 | | The FK1 domain of FKBP51 in complex with (1S,5S,6R)-10-((S)-3,5-dichloro-N-methylphenylsulfonimidoyl)-3-(pyridin-2-ylmethyl)-5-vinyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[S-[3,5-bis(chloranyl)phenyl]-N-methyl-sulfonimidoyl]-5-ethenyl-3-(pyridin-2-ylmethyl)-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Purder, P.L, Hausch, F. | | Deposit date: | 2023-02-08 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.01 Å) | | Cite: | Deconstructing Protein Binding of Sulfonamides and Sulfonamide Analogues.
Jacs Au, 3, 2023
|
|
5A70
 
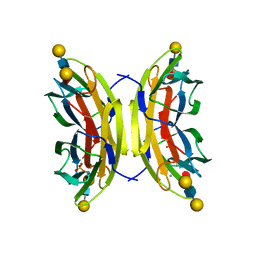 | | Structure of the LecB lectin from Pseudomonas aeruginosa strain PA14 in complex with lewis x tetrasaccharide | | Descriptor: | CALCIUM ION, LECB, SULFATE ION, ... | | Authors: | Sommer, R, Wagner, S, Varrot, A, Khaledi, A, Haussler, S, Imberty, A, Titz, A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Induction of rare conformation of oligosaccharide by binding to calcium-dependent bacterial lectin: X-ray crystallography and modelling study.
Eur.J.Med.Chem., 177, 2019
|
|
6VR8
 
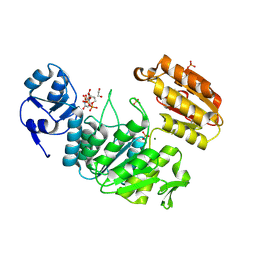 | | Structure of a pseudomurein peptide ligase type E from Methanothermus fervidus | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Mur ligase middle domain protein, ... | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | Deposit date: | 2020-02-06 | | Release date: | 2021-08-11 | | Last modified: | 2023-02-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterisation of methanogen pseudomurein cell wall peptide ligases homologous to bacterial MurE/F murein peptide ligases.
Microbiology (Reading, Engl.), 168, 2022
|
|
