7QQA
 
 | | MgADP-bound Fe protein of the iron-only nitrogenase from Azotobacter vinelandii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Trncik, C, Mueller, T, Franke, P, Einsle, O. | | Deposit date: | 2022-01-07 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | MgADP-bound Fe protein of the iron-only nitrogenase from Azotobacter vinelandii
Journal of Inorganic Biochemistry, 227, 2022
|
|
7BI7
 
 | | An Unexpected P-Cluster like Intermediate En Route to the Nitrogenase FeMo-co | | Descriptor: | FE(8)-S(7) CLUSTER, FeMo cofactor biosynthesis protein NifB, HYDROSULFURIC ACID, ... | | Authors: | Jenner, L.P, Cherrier, M.V, Amara, P, Rubio, L.M, Nicolet, Y. | | Deposit date: | 2021-01-12 | | Release date: | 2021-03-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | An unexpected P-cluster like intermediate en route to the nitrogenase FeMo-co.
Chem Sci, 12, 2021
|
|
3PDI
 
 | | Precursor bound NifEN | | Descriptor: | IRON/SULFUR CLUSTER, L-Cluster (Fe8S9), Nitrogenase MoFe cofactor biosynthesis protein NifE, ... | | Authors: | Kaiser, J.T, Hu, Y, Wiig, J.A, Rees, D.C, Ribbe, M.W. | | Deposit date: | 2010-10-22 | | Release date: | 2011-01-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of Precursor-Bound NifEN: A Nitrogenase FeMo Cofactor Maturase/Insertase.
Science, 331, 2011
|
|
3FWY
 
 | | Crystal structure of the L protein of Rhodobacter sphaeroides light-independent protochlorophyllide reductase (BchL) with MgADP bound: a homologue of the nitrogenase Fe protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase iron-sulfur ATP-binding protein, ... | | Authors: | Sarma, R, Barney, B.M, Hamilton, T.L, Jones, A, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2009-01-19 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal Structure of the L Protein of Rhodobacter sphaeroides Light-Independent Protochlorophyllide Reductase with MgADP Bound: A Homologue of the Nitrogenase Fe Protein.
Biochemistry, 47, 2008
|
|
8BOQ
 
 | | A. vinelandii Fe-nitrogenase FeFe protein | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE(8)-S(7) CLUSTER, Fe-only nitrogenase, ... | | Authors: | Trncik, C, Detemple, F, Einsle, O. | | Deposit date: | 2022-11-15 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.547 Å) | | Cite: | Iron-only Fe-nitrogenase underscores common catalytic principles in biological nitrogen fixation
Nat Catal, 2023
|
|
6OP1
 
 | |
7UT6
 
 | | C1 symmetric cryoEM structure of Azotobacter vinelandii MoFeP under non-turnover conditions | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Rutledge, H.L, Cook, B, Tezcan, F.A, Herzik, M.A. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.91 Å) | | Cite: | Structures of the nitrogenase complex prepared under catalytic turnover conditions.
Science, 377, 2022
|
|
7UT7
 
 | | C2 symmetric cryoEM structure of Azotobacter vinelandii MoFeP under non-turnover conditions | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Rutledge, H.L, Cook, B.D, Tezcan, F.A, Herzik, M.A. | | Deposit date: | 2022-04-26 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (1.91 Å) | | Cite: | Structures of the nitrogenase complex prepared under catalytic turnover conditions.
Science, 377, 2022
|
|
2KIC
 
 | | n-NafY. N-terminal domain of NafY | | Descriptor: | Nitrogenase gamma subunit | | Authors: | Phillips, A.H, Hernandez, J.A, Erbil, K, Pelton, J.G, Wemmer, D.E, Rubio, L.M. | | Deposit date: | 2009-05-01 | | Release date: | 2010-12-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Biological activity and solution structure of the apo-dinitrogenase binding domain of NafY
J.Biol.Chem., 2010
|
|
7MCI
 
 | | MoFe protein from Azotobacter vinelandii with a sulfur-replenished cofactor | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Kang, W, Lee, C, Hu, Y, Ribbe, M.W. | | Deposit date: | 2021-04-02 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Evidence of substrate binding and product release via belt-sulfur mobilization of the nitrogenase cofactor
Nat Catal, 5, 2022
|
|
6RK0
 
 | | Structure of the Flavocytochrome Anf3 from Azotobacter vinelandii | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Murray, J.W, Varghese, F, Kabasakal, B. | | Deposit date: | 2019-04-29 | | Release date: | 2019-05-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | A low-potential terminal oxidase associated with the iron-only nitrogenase from the nitrogen-fixing bacteriumAzotobacter vinelandii.
J.Biol.Chem., 294, 2019
|
|
5FFI
 
 | | [2Fe:2S] ferredoxin FeSII from Azotobacter vinelandii | | Descriptor: | Dimeric (2Fe-2S) protein, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Schlesier, J, Rohde, M, Gerhardt, S, Einsle, O. | | Deposit date: | 2015-12-18 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | A Conformational Switch Triggers Nitrogenase Protection from Oxygen Damage by Shethna Protein II (FeSII).
J.Am.Chem.Soc., 138, 2016
|
|
2YNM
 
 | | Structure of the ADPxAlF3-Stabilized Transition State of the Nitrogenase-like Dark-Operative Protochlorophyllide Oxidoreductase Complex from Prochlorococcus marinus with Its Substrate Protochlorophyllide a | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, ... | | Authors: | Krausze, J, Lange, C, Heinz, D.W, Moser, J. | | Deposit date: | 2012-10-16 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Adp-Aluminium Fluoride-Stabilized Protochlorophyllide Oxidoreductase Complex.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
2XDQ
 
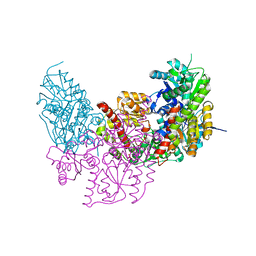 | | Dark Operative Protochlorophyllide Oxidoreductase (ChlN-ChlB)2 Complex | | Descriptor: | 1-METHYLGUANIDINE, IRON/SULFUR CLUSTER, LIGHT-INDEPENDENT PROTOCHLOROPHYLLIDE REDUCTASE SUBUNIT B, ... | | Authors: | Broecker, M.J, Schomburg, S, Heinz, D.W, Jahn, D, Schubert, W.-D, Moser, J. | | Deposit date: | 2010-05-06 | | Release date: | 2010-06-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Nitrogenase-Like Dark Operative Protochlorophyllide Oxidoreductase Catalytic Complex (Chln/Chlb)2.
J.Biol.Chem., 285, 2010
|
|
3O5T
 
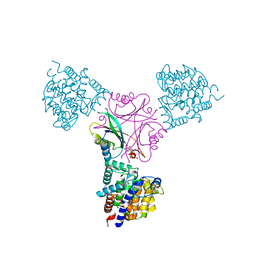 | | Structure of DraG-GlnZ complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Dinitrogenase reductase activacting glicohydrolase, MAGNESIUM ION, ... | | Authors: | Rajendran, C, Li, X.-D, Winkler, F.K. | | Deposit date: | 2010-07-28 | | Release date: | 2011-10-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of the GlnZ-DraG complex reveals a different form of PII-target interaction
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2WOD
 
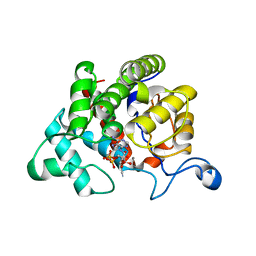 | | Crystal Structure of the dinitrogenase reductase-activating glycohydrolase (DRAG) from Rhodospirillum rubrum in complex with ADP- ribsoyllysine | | Descriptor: | ADP-RIBOSYL-[DINITROGEN REDUCTASE] GLYCOHYDROLASE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Berthold, C.L, Wang, H, Nordlund, S, Hogbom, M. | | Deposit date: | 2009-07-23 | | Release date: | 2009-08-11 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanism of Adp-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-Adp-Ribosylhydrolase Drag.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WOE
 
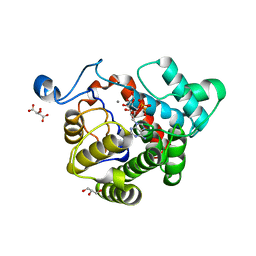 | | Crystal Structure of the D97N variant of dinitrogenase reductase- activating glycohydrolase (DRAG) from Rhodospirillum rubrum in complex with ADP-ribose | | Descriptor: | ADP-RIBOSYL-[DINITROGEN REDUCTASE] GLYCOHYDROLASE, GLYCEROL, L(+)-TARTARIC ACID, ... | | Authors: | Berthold, C.L, Wang, H, Nordlund, S, Hogbom, M. | | Deposit date: | 2009-07-23 | | Release date: | 2009-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of Adp-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-Adp-Ribosylhydrolase Drag.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WOC
 
 | | Crystal Structure of the dinitrogenase reductase-activating glycohydrolase (DRAG) from Rhodospirillum rubrum | | Descriptor: | ADP-RIBOSYL-[DINITROGEN REDUCTASE] GLYCOHYDROLASE, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Berthold, C.L, Wang, H, Nordlund, S, Hogbom, M. | | Deposit date: | 2009-07-23 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of Adp-Ribosylation Removal Revealed by the Structure and Ligand Complexes of the Dimanganese Mono-Adp-Ribosylhydrolase Drag.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3AET
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
2RE2
 
 | |
2KLA
 
 | | NMR STRUCTURE OF A PUTATIVE DINITROGENASE (MJ0327) FROM METHANOCOCCUS JANNASCHII | | Descriptor: | Uncharacterized protein MJ0327 | | Authors: | Jaudzems, K, Mohanty, B, Geralt, M, Serrano, P, Wilson, I, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2009-06-30 | | Release date: | 2009-08-11 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the protein NP_247299.1: comparison with the crystal structure.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2QTD
 
 | |
1O13
 
 | |
3AEQ
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N, ... | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
3AES
 
 | | Structure of the light-independent protochlorophyllide reductase catalyzing a key reduction for greening in the dark | | Descriptor: | IRON/SULFUR CLUSTER, Light-independent protochlorophyllide reductase subunit B, Light-independent protochlorophyllide reductase subunit N | | Authors: | Muraki, N, Nomata, J, Shiba, T, Fujita, Y, Kurisu, G. | | Deposit date: | 2010-02-10 | | Release date: | 2010-04-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-ray crystal structure of the light-independent protochlorophyllide reductase
Nature, 465, 2010
|
|
