7PRM
 
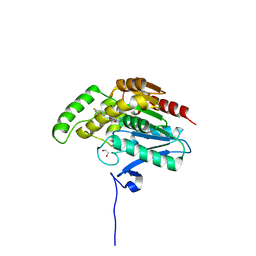 | | CRYSTAL STRUCTURE OF HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND 13 | | Descriptor: | (4~{R})-1-[4-(4-fluorophenyl)phenyl]-4-[4-(furan-2-ylcarbonyl)piperazin-1-yl]pyrrolidin-2-one, 1,2-ETHANEDIOL, Monoglyceride lipase | | Authors: | Grether, U, Gobbi, L, Kuhn, B, Collin, L, Leibrock, L, Heer, D, Wittwer, M, Benz, J. | | Deposit date: | 2021-09-22 | | Release date: | 2022-02-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Development of High Brain-Penetrant and Reversible Monoacylglycerol Lipase PET Tracers for Neuroimaging.
J.Med.Chem., 65, 2022
|
|
8IJO
 
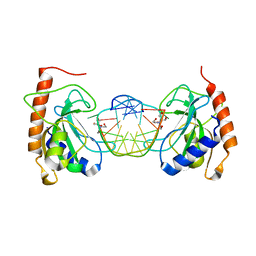 | |
8IK4
 
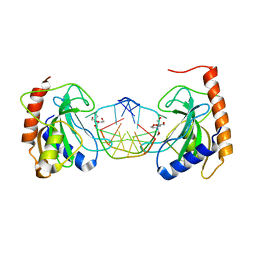 | |
8IJP
 
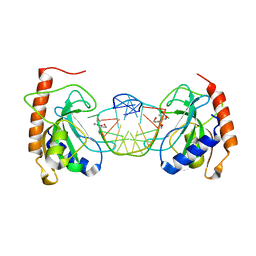 | |
8IK8
 
 | |
8IKD
 
 | |
8ILT
 
 | | Crystal structure of Est30 | | Descriptor: | Carboxylesterase | | Authors: | Feng, Y, Luo, Z. | | Deposit date: | 2023-03-04 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Crystal structure of Est30
To Be Published
|
|
2DJH
 
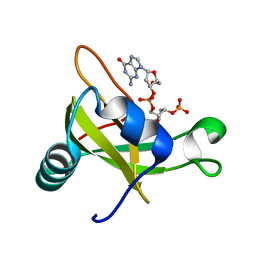 | | Crystal structure of the carboxy-terminal ribonuclease domain of Colicin E5 | | Descriptor: | 2'-DEOXYURIDINE 3'-MONOPHOSPHATE, 2-AMINO-9-(2-DEOXY-3-O-PHOSPHONOPENTOFURANOSYL)-1,9-DIHYDRO-6H-PURIN-6-ONE, Colicin-E5 | | Authors: | Yajima, S, Inoue, S, Ogawa, T, Nonaka, T, Ohsawa, K, Masaki, H. | | Deposit date: | 2006-04-03 | | Release date: | 2007-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for sequence-dependent recognition of colicin E5 tRNase by mimicking the mRNA-tRNA interaction
Nucleic Acids Res., 34, 2006
|
|
2ED6
 
 | |
2EQP
 
 | | Solution structure of the stn_TNFRSF12A_TNFR domain of Tumor necrosis factor receptor superfamily member 12A precursor | | Descriptor: | Tumor necrosis factor receptor superfamily member 12A | | Authors: | Dang, W, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2008-04-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the stn_TNFRSF12A_TNFR domain of Tumor necrosis factor receptor superfamily member 12A precursor
To be Published
|
|
2FCG
 
 | | Solution structure of the C-terminal fragment of human LL-37 | | Descriptor: | Antibacterial protein FALL-39, core peptide | | Authors: | Wang, G, Li, X. | | Deposit date: | 2005-12-12 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Human LL-37 Fragments and NMR-Based Identification of a Minimal Membrane-Targeting Antimicrobial and Anticancer Region.
J.Am.Chem.Soc., 128, 2006
|
|
8PTQ
 
 | | COMPLEX CRYSTAL STRUCTURE OF MUTANT HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND 5l | | Descriptor: | 1,2-ETHANEDIOL, Monoglyceride lipase, methyl 4-[(2~{S},3~{R})-3-(4-fluorophenyl)-1-(1-methanoylpiperidin-4-yl)-4-oxidanylidene-azetidin-2-yl]benzoate | | Authors: | Butini, S, Grether, U, Benz, J, Leibrock, L, Maramai, S, Papa, A, Carullo, G, Federico, S, Grillo, A, Di Guglielmo, B, Lamponi, S, Gemma, S, Campiani, G. | | Deposit date: | 2023-07-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of Potent and Selective Monoacylglycerol Lipase Inhibitors. SARs, Structural Analysis, and Biological Characterization.
J.Med.Chem., 67, 2024
|
|
8PTC
 
 | | COMPLEX CRYSTAL STRUCTURE OF MUTANT HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND 5d | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3~{R},4~{S})-2-oxidanylidene-3,4-diphenyl-azetidin-1-yl]piperidine-1-carbaldehyde, Monoglyceride lipase | | Authors: | Butini, S, Benz, J, Grether, U, Leibrock, L, Papa, A, Maramai, S, Carullo, G, Federico, S, Grillo, A, Di Guglielmo, B, Lamponi, S, Gemma, S, Campiani, G. | | Deposit date: | 2023-07-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Development of Potent and Selective Monoacylglycerol Lipase Inhibitors. SARs, Structural Analysis, and Biological Characterization.
J.Med.Chem., 67, 2024
|
|
8PTR
 
 | | COMPLEX CRYSTAL STRUCTURE OF MUTANT HUMAN MONOGLYCERIDE LIPASE WITH COMPOUND 5r | | Descriptor: | (3~{R},4~{S})-4-(1,3-benzodioxol-5-yl)-1-[1-(benzotriazol-1-ylcarbonyl)piperidin-4-yl]-3-(3-fluorophenyl)azetidin-2-one, 1,2-ETHANEDIOL, Monoglyceride lipase | | Authors: | Butini, S, Benz, J, Grether, U, Leibrock, L, Papa, A, Maramai, S, Carullo, G, Federico, S, Grillo, A, Di Guglielmo, B, Lamponi, S, Gemma, S, Campiani, G. | | Deposit date: | 2023-07-14 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Development of Potent and Selective Monoacylglycerol Lipase Inhibitors. SARs, Structural Analysis, and Biological Characterization.
J.Med.Chem., 67, 2024
|
|
2FBS
 
 | |
2FBU
 
 | | Solution structure of the N-terminal fragment of human LL-37 | | Descriptor: | Antibacterial protein FALL-39, core peptide | | Authors: | Wang, G, Li, X. | | Deposit date: | 2005-12-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Human LL-37 Fragments and NMR-Based Identification of a Minimal Membrane-Targeting Antimicrobial and Anticancer Region
J.Am.Chem.Soc., 128, 2006
|
|
2GOM
 
 | |
2EDM
 
 | |
4ILJ
 
 | |
5ZUN
 
 | | Crystal structure of human monoacylglycerol lipase in complex with compound 3l | | Descriptor: | (4R)-1-(2'-chloro[1,1'-biphenyl]-3-yl)-4-[4-(1,3-thiazole-2-carbonyl)piperazin-1-yl]pyrrolidin-2-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Sogabe, S, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2018-05-08 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design, Synthesis, and Evaluation of Piperazinyl Pyrrolidin-2-ones as a Novel Series of Reversible Monoacylglycerol Lipase Inhibitors
J. Med. Chem., 61, 2018
|
|
4JK7
 
 | | Open and closed forms of wild-type human PRP8 RNase H-like domain with bound Mg ion | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4JKD
 
 | | Open and closed forms of I1790Y human PRP8 RNase H-like domain with bound Mg ion | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Pre-mRNA-processing-splicing factor 8 | | Authors: | Schellenberg, M.J, Wu, T, Ritchie, D.B, Atta, K.A, MacMillan, A.M. | | Deposit date: | 2013-03-09 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A conformational switch in PRP8 mediates metal ion coordination that promotes pre-mRNA exon ligation.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4KE8
 
 | | Crystal structure of Monoglyceride lipase from Bacillus sp. H257 in complex with monopalmitoyl glycerol analogue | | Descriptor: | Thermostable monoacylglycerol lipase, tetradecyl hydrogen (R)-(3-azidopropyl)phosphonate | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
4KE6
 
 | | Crystal structure D196N mutant of Monoglyceride lipase from Bacillus sp. H257 in complex with 1-rac-lauroyl glycerol | | Descriptor: | (2R)-2,3-dihydroxypropyl dodecanoate, (4S)-2-METHYL-2,4-PENTANEDIOL, Thermostable monoacylglycerol lipase | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
4KE7
 
 | | Crystal structure of Monoglyceride lipase from Bacillus sp. H257 in complex with an 1-myristoyl glycerol analogue | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Thermostable monoacylglycerol lipase, dodecyl hydrogen (S)-(3-azidopropyl)phosphonate | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2013-11-20 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
