6GVS
 
 | | Engineered glycolyl-CoA reductase comprising 8 mutations with bound NADP+ | | Descriptor: | Aldehyde dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION | | Authors: | Zarzycki, J, Trudeau, D, Scheffen, M, Erb, T.J, Tawfik, D.S. | | Deposit date: | 2018-06-21 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.579 Å) | | Cite: | Design and in vitro realization of carbon-conserving photorespiration.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6H2F
 
 | | Structure of the pre-pore AhlB of the tripartite alpha-pore forming toxin, AHL, from Aeromonas hydrophila. | | Descriptor: | AhlB, PHOSPHATE ION | | Authors: | Churchill-Angus, A.M, Wilson, J.S, Baker, P.J. | | Deposit date: | 2018-07-13 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Identification and structural analysis of the tripartite alpha-pore forming toxin of Aeromonas hydrophila.
Nat Commun, 10, 2019
|
|
8DJ4
 
 | | NMR Solution Structure of C-terminally amidated, Full-length Human Galanin | | Descriptor: | Galanin | | Authors: | Wilkinson, R.E, Kraichely, K.N, Buchanan, L.E, Parnham, S, Giuliano, M.W. | | Deposit date: | 2022-06-30 | | Release date: | 2022-08-24 | | Last modified: | 2022-09-07 | | Method: | SOLUTION NMR | | Cite: | The neuropeptide galanin adopts an irregular secondary structure.
Biochem.Biophys.Res.Commun., 626, 2022
|
|
3SKG
 
 | | Crystal structure of beta-site app-cleaving enzyme 1 (BACE-WT) complex with (2S)-2-((3R)-3-acetamido-3-isobutyl-2-oxo-1-pyrrolidinyl)-N-((1S,2R)-1-(3,5-difluorobenzyl)-2-hydroxy-2-(1,2,3,4-tetrahydro-3-isoquinolinyl)ethyl)-4-phenylbutanamide | | Descriptor: | (2S)-2-[(3R)-3-(acetylamino)-3-(2-methylpropyl)-2-oxopyrrolidin-1-yl]-N-{(1R,2S)-3-(3,5-difluorophenyl)-1-hydroxy-1-[(3R)-1,2,3,4-tetrahydroisoquinolin-3-yl]propan-2-yl}-4-phenylbutanamide, Beta-secretase 1 | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2011-06-22 | | Release date: | 2011-09-07 | | Last modified: | 2012-12-05 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Synthesis and in vivo evaluation of cyclic diaminopropane BACE-1 inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
1RYX
 
 | |
5KOX
 
 | |
7O3C
 
 | | Murine supercomplex CIII2CIV in the mature unlocked conformation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and assembly of the mammalian mitochondrial supercomplex CIII 2 CIV.
Nature, 598, 2021
|
|
7O3E
 
 | | Murine supercomplex CIII2CIV in the intermediate locked conformation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and assembly of the mammalian mitochondrial supercomplex CIII 2 CIV.
Nature, 598, 2021
|
|
7O37
 
 | | Murine supercomplex CIII2CIV in the assembled locked conformation | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure and assembly of the mammalian mitochondrial supercomplex CIII 2 CIV.
Nature, 598, 2021
|
|
7O3H
 
 | | Murine CIII2 focus-refined from supercomplex CICIII2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2021-04-01 | | Release date: | 2021-10-13 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structure and assembly of the mammalian mitochondrial supercomplex CIII 2 CIV.
Nature, 598, 2021
|
|
6YDJ
 
 | | P146A variant of beta-phosphoglucomutase from Lactococcus lactis in complex with glucose 6-phosphate and trifluoromagnesate | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, 6-O-phosphono-beta-D-glucopyranose, ... | | Authors: | Wood, H.P, Cruz-Navarrete, F.A, Baxter, N.J, Trevitt, C.R, Robertson, A.J, Dix, S.R, Hounslow, A.M, Cliff, M.J, Waltho, J.P. | | Deposit date: | 2020-03-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Allomorphy as a mechanism of post-translational control of enzyme activity.
Nat Commun, 11, 2020
|
|
1S3P
 
 | |
6YDK
 
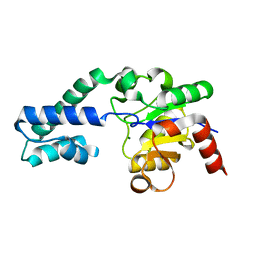 | | Substrate-free P146A variant of beta-phosphoglucomutase from Lactococcus lactis | | Descriptor: | Beta-phosphoglucomutase, MAGNESIUM ION | | Authors: | Wood, H.P, Cruz-Navarrete, F.A, Baxter, N.J, Trevitt, C.R, Robertson, A.J, Dix, S.R, Hounslow, A.M, Cliff, M.J, Waltho, J.P. | | Deposit date: | 2020-03-20 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Allomorphy as a mechanism of post-translational control of enzyme activity.
Nat Commun, 11, 2020
|
|
3R6H
 
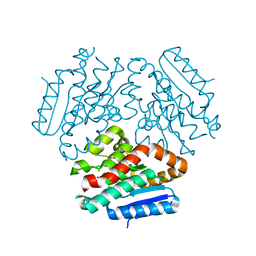 | |
1OI9
 
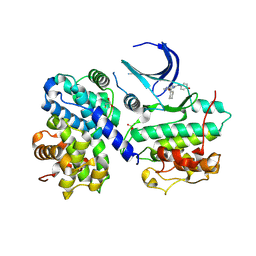 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with a 6-cyclohexylmethyloxy-2-anilino-purine inhibitor | | Descriptor: | 6-CYCLOHEXYLMETHYLOXY-2-(4'-HYDROXYANILINO)PURINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2003-06-10 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | N2-Substituted O6-Cyclohexylmethylguanine Derivatives: Potent Inhibitors of Cyclin-Dependent Kinases 1 and 2
J.Med.Chem., 47, 2004
|
|
3RD5
 
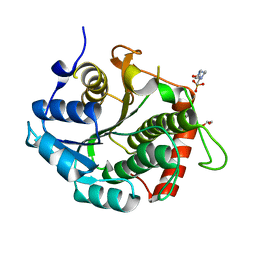 | |
1DWN
 
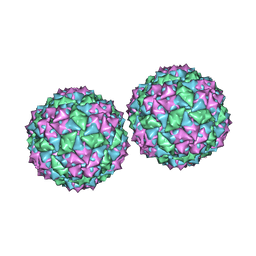 | | Structure of bacteriophage PP7 from Pseudomonas aeruginosa at 3.7 A resolution | | Descriptor: | PHAGE COAT PROTEIN | | Authors: | Tars, K, Fridborg, K, Bundule, M, Liljas, L. | | Deposit date: | 1999-12-09 | | Release date: | 2000-02-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure Determination of Phage Pp7 from Pseudomonas Aeruginosa: From Poor Data to a Good Map
Acta Crystallogr.,Sect.D, 56, 2000
|
|
1OIU
 
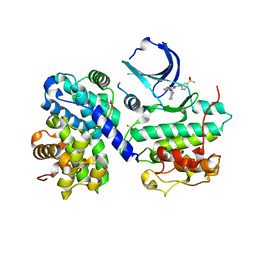 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with a 6-cyclohexylmethyloxy-2-anilino-purine inhibitor | | Descriptor: | 3-(6-CYCLOHEXYLMETHOXY-9H-PURIN-2-YLAMINO)-BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2003-06-26 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N2-Substituted O6-Cyclohexylmethylguanine Derivatives: Potent Inhibitors of Cyclin-Dependent Kinases 1 and 2
J.Med.Chem., 47, 2004
|
|
1U4N
 
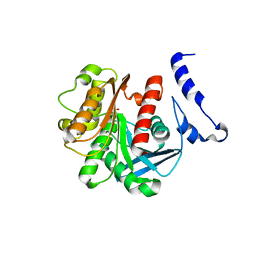 | | Crystal Structure Analysis of the M211S/R215L EST2 mutant | | Descriptor: | CARBOXYLESTERASE EST2, SULFATE ION | | Authors: | De Simone, G, Menchise, V, Alterio, V, Mandrich, L, Rossi, M, Manco, G, Pedone, C. | | Deposit date: | 2004-07-26 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Crystal Structure of an EST2 Mutant Unveils Structural Insights on the H Group of the Carboxylesterase/Lipase Family.
J.Mol.Biol., 343, 2004
|
|
1OIQ
 
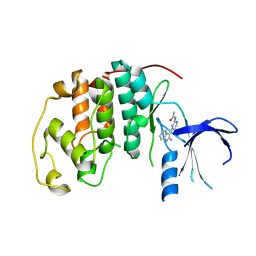 | | Imidazopyridines: a potent and selective class of Cyclin-dependent Kinase inhibitors identified through Structure-based hybridisation | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-[4-(2-METHYLIMIDAZO[1,2-A]PYRIDIN-3-YL)-2-PYRIMIDINYL]ACETAMIDE | | Authors: | Beattie, J.F, Breault, G.A, Byth, K.F, Culshaw, J.D, Ellston, R.P.A, Green, S, Minshull, C.A, Norman, R.A, Pauptit, R.A, Thomas, A.P, Jewsbury, P.J. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Imidazo[1,2-A]Pyridines: A Potent and Selective Class of Cyclin-Dependent Kinase Inhibitors Identified Through Structure-Based Hybridisation
Bioorg.Med.Chem.Lett., 13, 2003
|
|
5KZQ
 
 | | Metabotropic Glutamate Receptor in complex with antagonist (1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-azanyl-3-[[3,4-bis(fluoranyl)phenyl]sulfanylmethyl]-4-oxidanyl-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid | | Descriptor: | (1~{S},2~{R},3~{S},4~{S},5~{R},6~{R})-2-azanyl-3-[[3,4-bis(fluoranyl)phenyl]sulfanylmethyl]-4-oxidanyl-bicyclo[3.1.0]hexane-2,6-dicarboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Metabotropic glutamate receptor 2 | | Authors: | Chappell, M.D, Li, R, Smith, S.C, Dressman, B.A, Tromiczak, E.G, Tripp, A.E, Blanco, M.-J, Vetman, T, Quimby, S.J, Matt, J, Britton, T, Fivush, A.M, Schkeryantz, J.M, Mayhugh, D, Erickson, J.A, Bures, M, Jaramillo, C, Carpintero, M, de Diego, J.E, Barberis, M, Garcia-Cerrada, S, Soriano, J.F, Antonysamy, S, Atwell, S, MacEwan, I, Condon, B, Bradley, C, Wang, J, Zhang, A, Conners, K, Groshong, C, Wasserman, S.R, Koss, J.W, Witkin, J.M, Li, X, Overshiner, C, Wafford, K.A, Seidel, W, Wang, X.-S, Heinz, B.A, Swanson, S, Catlow, J, Bedwell, D, Monn, J.A, Mitch, C.H, Ornstein, P. | | Deposit date: | 2016-07-25 | | Release date: | 2016-12-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of (1S,2R,3S,4S,5R,6R)-2-Amino-3-[(3,4-difluorophenyl)sulfanylmethyl]-4-hydroxy-bicyclo[3.1.0]hexane-2,6-dicarboxylic Acid Hydrochloride (LY3020371HCl): A Potent, Metabotropic Glutamate 2/3 Receptor Antagonist with Antidepressant-Like Activity.
J. Med. Chem., 59, 2016
|
|
1O8D
 
 | |
1O8M
 
 | |
1O8E
 
 | |
1O8L
 
 | |
