7OJ0
 
 | | Cryo-EM structure of 70S ribosome stalled with TnaC peptide and RF2 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Su, T, Kudva, R, Becker, T, Berninghausen, O, Heijne, G, Cheng, J, Beckmann, R. | | Deposit date: | 2021-05-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of l-tryptophan-dependent inhibition of release factor 2 by the TnaC arrest peptide.
Nucleic Acids Res., 49, 2021
|
|
7OIZ
 
 | | Cryo-EM structure of 70S ribosome stalled with TnaC peptide | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Su, T, Kudva, R, Becker, T, Berninghausen, O, Heijne, G, Cheng, J, Beckmann, R. | | Deposit date: | 2021-05-13 | | Release date: | 2021-09-15 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of l-tryptophan-dependent inhibition of release factor 2 by the TnaC arrest peptide.
Nucleic Acids Res., 49, 2021
|
|
1STQ
 
 | | Cyrstal Structure of Cytochrome c Peroxidase Mutant: CcPK2M3 | | Descriptor: | Cytochrome c peroxidase, mitochondrial, POTASSIUM ION, ... | | Authors: | Barrows, T.P, Bhaskar, B, Poulos, T.L. | | Deposit date: | 2004-03-25 | | Release date: | 2004-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Electrostatic control of the tryptophan radical in cytochrome c peroxidase.
Biochemistry, 43, 2004
|
|
5KSF
 
 | |
3ZJI
 
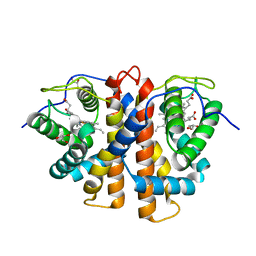 | | Tyr(61)B10Ala mutation of M.acetivorans protoglobin in complex with cyanide | | Descriptor: | CYANIDE ION, GLYCEROL, PROTOGLOBIN, ... | | Authors: | Pesce, A, Tilleman, L, Donne, J, Aste, E, Ascenzi, P, Ciaccio, C, Coletta, M, Moens, L, Viappiani, C, Dewilde, S, Bolognesi, M, Nardini, M. | | Deposit date: | 2013-01-18 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and Haem-Distal Site Plasticity in Methanosarcina Acetivorans Protoglobin.
Plos One, 8, 2013
|
|
5V4O
 
 | |
5KQ6
 
 | |
5KQH
 
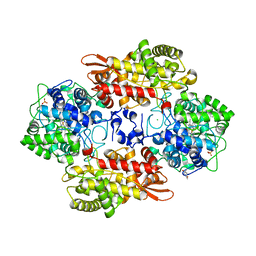 | |
5KQ3
 
 | |
5KQ0
 
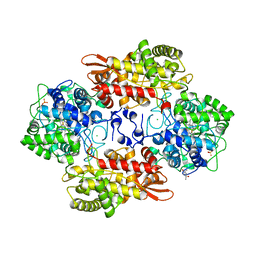 | |
3BK9
 
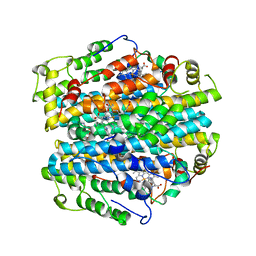 | | H55A mutant of tryptophan 2,3-dioxygenase from Xanthomonas campestris | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, TRYPTOPHAN, Tryptophan 2,3-dioxygenase | | Authors: | Bruckmann, C, Mowat, C.G. | | Deposit date: | 2007-12-06 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling substrate binding
Biochemistry, 47, 2008
|
|
2VYZ
 
 | | Mutant Ala55Phe of Cerebratulus lacteus mini-hemoglobin | | Descriptor: | ACETATE ION, GLYCEROL, NEURAL HEMOGLOBIN, ... | | Authors: | Salter, M.D, Nienhaus, K, Nienhaus, G.U, Dewilde, S, Moens, L, Pesce, A, Nardini, M, Bolognesi, M, Olson, J.S. | | Deposit date: | 2008-07-29 | | Release date: | 2008-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Apolar Channel in Cerebratulus Lacteus Hemoglobin is the Route for O2 Entry and Exit.
J.Biol.Chem., 283, 2008
|
|
3EP2
 
 | | Model of Phe-tRNA(Phe) in the ribosomal pre-accommodated state revealed by cryo-EM | | Descriptor: | 30S ribosomal protein S12, 50S ribosomal protein L11, Elongation factor Tu, ... | | Authors: | Frank, J, Li, W, Agirrezabala, X. | | Deposit date: | 2008-09-29 | | Release date: | 2008-12-16 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Recognition of aminoacyl-tRNA: a common molecular mechanism revealed by cryo-EM.
Embo J., 27, 2008
|
|
3E08
 
 | | H55S mutant Xanthomonas campestris tryptophan 2,3-dioxygenase | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, TRYPTOPHAN, Tryptophan 2,3-dioxygenase | | Authors: | Mowat, C.G, Campbell, L.P. | | Deposit date: | 2008-07-31 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Histidine 55 of tryptophan 2,3-dioxygenase is not an active site base but regulates catalysis by controlling substrate binding
Biochemistry, 47, 2008
|
|
1SWT
 
 | | CORE-STREPTAVIDIN MUTANT D128A IN COMPLEX WITH BIOTIN AT PH 4.5 | | Descriptor: | BIOTIN, PROTEIN (STREPTAVIDIN) | | Authors: | Freitag, S, Le Trong, I, Chu, V, Klumb, L.A, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-10-22 | | Release date: | 1999-07-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural snapshot of an intermediate on the streptavidin-biotin dissociation pathway.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1SWS
 
 | | CORE-STREPTAVIDIN MUTANT D128A AT PH 4.5 | | Descriptor: | PROTEIN (STREPTAVIDIN) | | Authors: | Freitag, S, Chu, V, Le Trong, I, Klumb, L.A, To, R, Stayton, P.S, Stenkamp, R.E. | | Deposit date: | 1998-10-22 | | Release date: | 1999-07-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural snapshot of an intermediate on the streptavidin-biotin dissociation pathway.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
4ICS
 
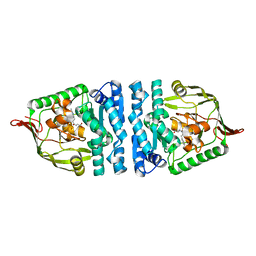 | | Crystal structure of PepS from Streptococcus pneumoniae in complex with a substrate | | Descriptor: | Aminopeptidase PepS, GLYCINE, TRYPTOPHAN, ... | | Authors: | Lee, S, Kim, K.K, Ta, M.H. | | Deposit date: | 2012-12-11 | | Release date: | 2013-10-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structure-based elucidation of the regulatory mechanism for aminopeptidase activity.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1K7E
 
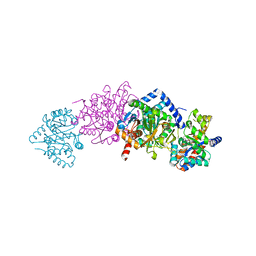 | | CRYSTAL STRUCTURE OF WILD-TYPE TRYPTOPHAN SYNTHASE COMPLEXED WITH N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID | | Descriptor: | N-[1H-INDOL-3-YL-ACETYL]GLYCINE ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Weyand, M, Schlichting, I, Marabotti, A, Mozzarelli, A. | | Deposit date: | 2001-10-19 | | Release date: | 2002-07-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of a new class of allosteric effectors complexed to tryptophan synthase.
J.Biol.Chem., 277, 2002
|
|
6OS5
 
 | |
3NMM
 
 | | Human Hemoglobin A mutant alpha H58W deoxy-form | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Birukou, I, Soman, J, Olson, J.S. | | Deposit date: | 2010-06-22 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Blocking the gate to ligand entry in human hemoglobin.
J.Biol.Chem., 286, 2011
|
|
3NML
 
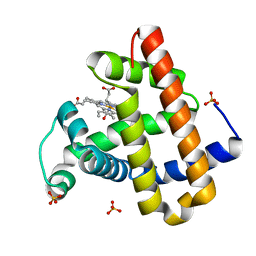 | | Sperm whale myoglobin mutant H64W carbonmonoxy-form | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Birukou, I, Soman, J, Olson, J.S. | | Deposit date: | 2010-06-22 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Blocking the gate to ligand entry in human hemoglobin.
J.Biol.Chem., 286, 2011
|
|
3OGB
 
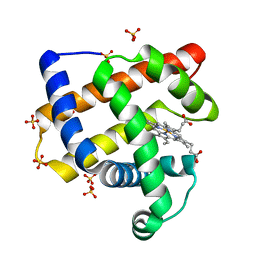 | |
6XAM
 
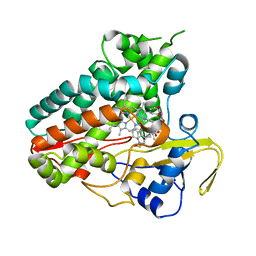 | | Crystal structure of NzeB in complex with cyclo-(L-Trp-L-homoalanine) | | Descriptor: | (3S,6S)-3-ethyl-6-[(1H-indol-3-yl)methyl]piperazine-2,5-dione, 1,2-ETHANEDIOL, NzeB, ... | | Authors: | Shende, V.V, Khatri, Y, Newmister, S.A, Sanders, J.N, Lindovska, P, Yu, F, Doyon, T.J, Kim, J, Movassaghi, M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-04 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.481 Å) | | Cite: | Structure and Function of NzeB, a Versatile C-C and C-N Bond-Forming Diketopiperazine Dimerase.
J.Am.Chem.Soc., 142, 2020
|
|
4V6K
 
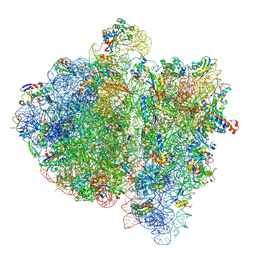 | | Structural insights into cognate vs. near-cognate discrimination during decoding. | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Agirrezabala, X, Schreiner, E, Trabuco, L.G, Lei, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-01-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.25 Å) | | Cite: | Structural insights into cognate versus near-cognate discrimination during decoding.
Embo J., 30, 2011
|
|
4V6L
 
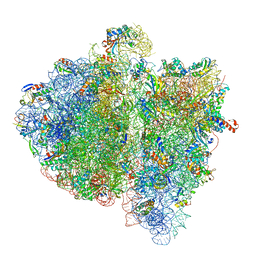 | | Structural insights into cognate vs. near-cognate discrimination during decoding. | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Agirrezabala, X, Schreiner, E, Trabuco, L.G, Lei, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-01-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (13.2 Å) | | Cite: | Structural insights into cognate versus near-cognate discrimination during decoding.
Embo J., 30, 2011
|
|
