2B5Q
 
 | |
3SB2
 
 | | Crystal Structure of the RNA chaperone Hfq from Herbaspirillum seropedicae SMR1 | | Descriptor: | GLYCEROL, Protein hfq | | Authors: | Kadowaki, M.A.S, Iulek, J, Barbosa, J.A.R.G, Pedrosa, F.O, Souza, E.M, Chubatsu, L.S, Monteiro, R.A, Steffens, M.B.R. | | Deposit date: | 2011-06-03 | | Release date: | 2012-01-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6301 Å) | | Cite: | Structural characterization of the RNA chaperone Hfq from the nitrogen-fixing bacterium Herbaspirillum seropedicae SmR1.
Biochim.Biophys.Acta, 1824, 2011
|
|
3K3E
 
 | | Crystal structure of the PDE9A catalytic domain in complex with (R)-BAY73-6691 | | Descriptor: | 1-(2-chlorophenyl)-6-[(2R)-3,3,3-trifluoro-2-methylpropyl]-1,7-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Wang, H, Luo, X, Ye, M, Hou, J, Robinson, H, Ke, H. | | Deposit date: | 2009-10-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insight into Binding of Phosphodiesterase-9A Selective Inhibitors by Crystal Structures and Mutagenesis
J.Med.Chem., 53, 2010
|
|
3F5U
 
 | | Crystal structure of the death associated protein kinase in complex with AMPPNP and Mg2+ | | Descriptor: | Death-associated protein kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | McNamara, L.K, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2008-11-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into nucleotide recognition by human death-associated protein kinase.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3F9Q
 
 | |
7E0M
 
 | | Crystal structure of phospholipase D | | Descriptor: | Phospholipase, SULFATE ION | | Authors: | Wang, F.H. | | Deposit date: | 2021-01-28 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of a Phospholipase D from the Plant-Associated Bacteria Serratia plymuthica Strain AS9 Reveals a Unique Arrangement of Catalytic Pocket.
Int J Mol Sci, 22, 2021
|
|
2PRC
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOPSEUDOMONAS VIRIDIS (UBIQUINONE-2 COMPLEX) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Lancaster, C.R.D, Michel, H. | | Deposit date: | 1997-07-29 | | Release date: | 1998-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The coupling of light-induced electron transfer and proton uptake as derived from crystal structures of reaction centres from Rhodopseudomonas viridis modified at the binding site of the secondary quinone, QB.
Structure, 5, 1997
|
|
2RB5
 
 | |
3HPF
 
 | | Crystal structure of the mutant Y90F of divergent galactarate dehydratase from Oceanobacillus iheyensis complexed with Mg and galactarate | | Descriptor: | D-galactaric acid, MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-06-04 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
2B5P
 
 | |
4ARU
 
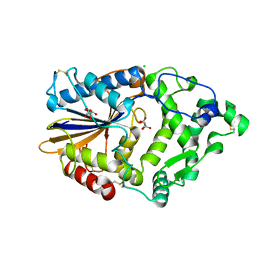 | | Hafnia Alvei phytase in complex with tartrate | | Descriptor: | CHLORIDE ION, HISTIDINE ACID PHOSPHATASE, L(+)-TARTARIC ACID, ... | | Authors: | Ariza, A, Moroz, O.V, Blagova, E.B, Turkenburg, J.P, Vevodova, J, Roberts, S, Vind, J, Sjoholm, C, Lassen, S.F, De Maria, L, Glitsoe, V, Skov, L.K, Wilson, K.S. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Degradation of Phytate by the 6-Phytase from Hafnia Alvei: A Combined Structural and Solution Study.
Plos One, 8, 2013
|
|
2KPY
 
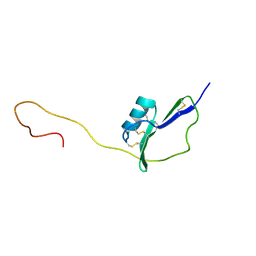 | | Solution Structure of the major allergen of Artemisia vulgaris (Art v 1) | | Descriptor: | Major pollen allergen Art v 1 | | Authors: | Razzera, G, Gadermaier, G, Almeida, M, Egger, M, Jahn-Schmid, B, Almeida, F, Ferreira, F, Valente, A. | | Deposit date: | 2009-10-23 | | Release date: | 2010-10-06 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Mapping the interactions between a major pollen allergen and human IgE antibodies.
Structure, 18, 2010
|
|
2RBK
 
 | |
3GM7
 
 | |
4ARS
 
 | | Hafnia Alvei phytase apo form | | Descriptor: | ACETATE ION, GLYCEROL, HISTIDINE ACID PHOSPHATASE | | Authors: | Ariza, A, Moroz, O.V, Blagova, E.B, Turkenburg, J.P, Vevodova, J, Roberts, S, Vind, J, Sjoholm, C, Lassen, S.F, De Maria, L, Glitsoe, V, Skov, L.K, Wilson, K.S. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Degradation of Phytate by the 6-Phytase from Hafnia Alvei: A Combined Structural and Solution Study.
Plos One, 8, 2013
|
|
4MYL
 
 | |
3QI3
 
 | | Crystal structure of PDE9A(Q453E) in complex with inhibitor BAY73-6691 | | Descriptor: | 1-(2-chlorophenyl)-6-[(2R)-3,3,3-trifluoro-2-methylpropyl]-1,7-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Hou, J, Xu, J, Liu, M, Zhao, R, Lou, H, Ke, H. | | Deposit date: | 2011-01-26 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural asymmetry of phosphodiesterase-9, potential protonation of a glutamic Acid, and role of the invariant glutamine.
Plos One, 6, 2011
|
|
4MYF
 
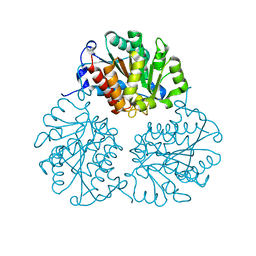 | | Crystal structure of Trypanosoma cruzi formiminoglutamase(oxidized) with Mn2+2 at pH 6.0 | | Descriptor: | Formiminoglutamase, MANGANESE (II) ION | | Authors: | Hai, Y, Dugery, R.J, Healy, D, Christianson, D.W. | | Deposit date: | 2013-09-27 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Formiminoglutamase from trypanosoma cruzi is an arginase-like manganese metalloenzyme.
Biochemistry, 52, 2013
|
|
4MYK
 
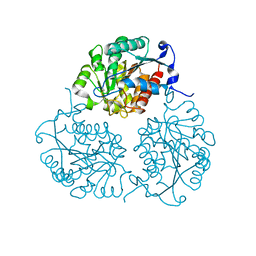 | | Crystal structure of Trypanosoma cruzi formiminoglutamase (oxidized) with Mn2+2 at pH 8.5 | | Descriptor: | Formiminoglutamase, MANGANESE (II) ION | | Authors: | Hai, Y, Dugery, R.J, Healy, D, Christianson, D.W. | | Deposit date: | 2013-09-27 | | Release date: | 2013-11-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.518 Å) | | Cite: | Formiminoglutamase from trypanosoma cruzi is an arginase-like manganese metalloenzyme.
Biochemistry, 52, 2013
|
|
4MXR
 
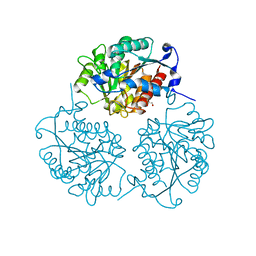 | | Crystal structure of Trypanosoma cruzi formiminoglutamase with Mn2+2 | | Descriptor: | Formiminoglutamase, GLYCEROL, MANGANESE (II) ION | | Authors: | Hai, Y, Dugery, R.-J, Healy, D, Christianson, D.W. | | Deposit date: | 2013-09-26 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Formiminoglutamase from trypanosoma cruzi is an arginase-like manganese metalloenzyme.
Biochemistry, 52, 2013
|
|
4ARV
 
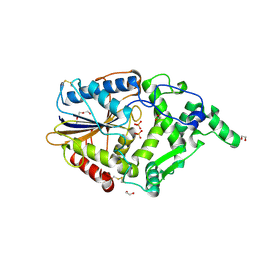 | | Yersinia kristensenii phytase apo form | | Descriptor: | 1,2-ETHANEDIOL, 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ... | | Authors: | Ariza, A, Moroz, O.V, Blagova, E.B, Turkenburg, J.P, Vevodova, J, Roberts, S, Vind, J, Sjoholm, C, Lassen, S.F, De Maria, L, Glitsoe, V, Skov, L.K, Wilson, K.S. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Degradation of Phytate by the 6-Phytase from Hafnia Alvei: A Combined Structural and Solution Study.
Plos One, 8, 2013
|
|
4ARO
 
 | | Hafnia Alvei phytase in complex with myo-inositol hexakis sulphate | | Descriptor: | D-MYO-INOSITOL-HEXASULPHATE, DI(HYDROXYETHYL)ETHER, HISTIDINE ACID PHOSPHATASE, ... | | Authors: | Moroz, O.V, Blagova, E.B, Ariza, A, Turkenburg, J.P, Vevodova, J, Roberts, S, Vind, J, Sjoholm, C, Lassen, S.F, De Maria, L, Glitsoe, V, Skov, L.K, Wilson, K.S. | | Deposit date: | 2012-04-25 | | Release date: | 2013-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Degradation of Phytate by the 6-Phytase from Hafnia Alvei: A Combined Structural and Solution Study.
Plos One, 8, 2013
|
|
2IN2
 
 | |
2EIR
 
 | |
2KMU
 
 | |
