8X30
 
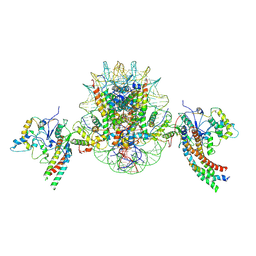 | | Structure of piccolo NuA4 and H2A.Z nucleosome 2:1 complex | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X2Y
 
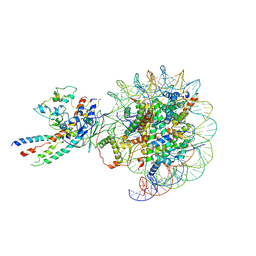 | | The class1 of piccolo NuA4 bound to the H2A.Z nucleosome complex at harboring state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X2X
 
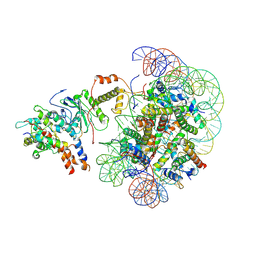 | | The piccolo NuA4 bound to the H2A.Z nucleosome complex at pre-H4-acetylation state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8X31
 
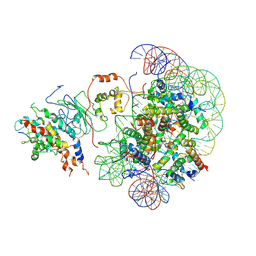 | | The piccolo NuA4 bound to the H2A.Z nucleosome complex with Ac-CoA at resetting state | | Descriptor: | Chromatin modification-related protein, DNA (146-MER), Histone H2A, ... | | Authors: | Wang, L, Zhang, H, Zhu, H, Zhu, P. | | Deposit date: | 2023-11-10 | | Release date: | 2025-03-19 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM structures reveal the acetylation process of piccolo NuA4.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
8PP7
 
 | | human RYBP-PRC1 bound to mononucleosome | | Descriptor: | DNA (215-mer), E3 ubiquitin-protein ligase RING2, Histone H2A, ... | | Authors: | Ciapponi, M, Benda, C, Mueller, J. | | Deposit date: | 2023-07-06 | | Release date: | 2024-03-27 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis of the histone ubiquitination read-write mechanism of RYBP-PRC1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8PP6
 
 | | human RYBP-PRC1 bound to H2AK118ub1 nucleosome | | Descriptor: | DNA (215-MER), Histone H2A, Histone H2B, ... | | Authors: | Ciapponi, M, Benda, C, Mueller, J. | | Deposit date: | 2023-07-06 | | Release date: | 2024-04-03 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural basis of the histone ubiquitination read-write mechanism of RYBP-PRC1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8Q15
 
 | | CryoEM structure of canonical rice nucleosome core particle | | Descriptor: | Histone H2A.2, Histone H2B.4, Histone H3.2, ... | | Authors: | Sotelo-Parrilla, P, Hari Sundar G, V, Raju, S, Jha, S, Gireesh, A, Gut, F, Vinothkumar, K.R, Berger, F, Shivaprasad, P.V, Jeyaprakash, A.A. | | Deposit date: | 2023-07-30 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An Oryza-specific histone H4 variant predisposes H4 lysine 5 acetylation to modulate salt stress responses.
Nat.Plants, 11, 2025
|
|
8Q16
 
 | | CryoEM structure of rice nucleosome containing a H4 variant chimera | | Descriptor: | Chimeric H4.V, Histone H2A.2, Histone H2B.4, ... | | Authors: | Sotelo-Parrilla, P, Hari Sundar G, V, Raju, S, Jha, S, Gireesh, A, Gut, F, Vinothkumar, K.R, Berger, F, Shivaprasad, P.V, Jeyaprakash, A.A. | | Deposit date: | 2023-07-30 | | Release date: | 2025-02-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | An Oryza-specific histone H4 variant predisposes H4 lysine 5 acetylation to modulate salt stress responses.
Nat.Plants, 11, 2025
|
|
8EUJ
 
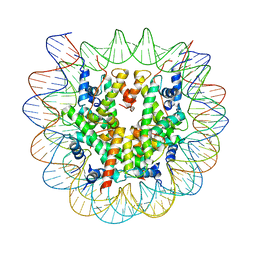 | | Class2 of the INO80-Nucleosome complex | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | Deposit date: | 2022-10-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETT
 
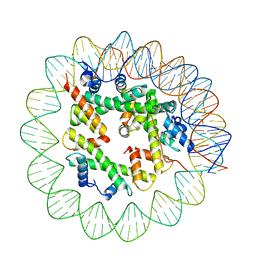 | | Class1 of the INO80-Hexasome complex | | Descriptor: | DNA (110-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-12 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (6.68 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8ETV
 
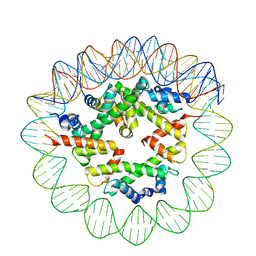 | | Class2 of the INO80-Hexasome complex | | Descriptor: | DNA (110-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-17 | | Release date: | 2023-07-12 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8EU2
 
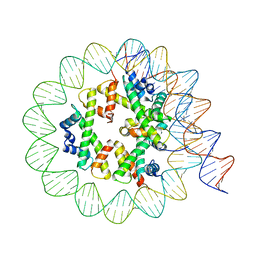 | | Class3 of the INO80-Hexasome complex | | Descriptor: | DNA (110-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Cheng, Y.F, Narlikar, G. | | Deposit date: | 2022-10-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
8EUE
 
 | | Class1 of the INO80-Nucleosome complex | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Wu, H, Munoz, E, Gourdet, M, Narlikar, G, Cheng, Y.F. | | Deposit date: | 2022-10-18 | | Release date: | 2023-07-12 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Reorientation of INO80 on hexasomes reveals basis for mechanistic versatility.
Science, 381, 2023
|
|
7WBV
 
 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-4) of the nucleosome | | Descriptor: | DNA (159-MER), DNA (198-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2021-12-17 | | Release date: | 2023-07-05 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
7WBX
 
 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-3) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2021-12-17 | | Release date: | 2023-07-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
7WBW
 
 | | RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-3.5) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Osumi, K, Kujirai, T, Ehara, H, Sekine, S, Takizawa, Y, Kurumizaka, H. | | Deposit date: | 2021-12-17 | | Release date: | 2023-07-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structural Basis of Damaged Nucleotide Recognition by Transcribing RNA Polymerase II in the Nucleosome.
J.Mol.Biol., 435, 2023
|
|
8QZM
 
 | | Structure of DNMT3A1 UDR region bound to H2AK119ub nucleosome | | Descriptor: | DNA (145-MER), DNA (cytosine-5)-methyltransferase 3A, Histone H2A type 1, ... | | Authors: | Burdett, H, Wapenaar, H, Wilson, M.D. | | Deposit date: | 2023-10-27 | | Release date: | 2024-10-23 | | Last modified: | 2024-12-18 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The N-terminal region of DNMT3A engages the nucleosome surface to aid chromatin recruitment.
Embo Rep., 25, 2024
|
|
7XSE
 
 | | RNA polymerase II elongation complex transcribing a nucleosome (EC42) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-13 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XSX
 
 | | RNA polymerase II elongation complex transcribing a nucleosome (EC49) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-15 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XTD
 
 | | RNA polymerase II elongation complex transcribing a nucleosome (EC58oct) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-16 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
7XSZ
 
 | | RNA polymerase II elongation complex transcribing a nucleosome (EC115) | | Descriptor: | Component of the Paf1p complex, Constituent of Paf1 complex with RNA polymerase II, Paf1p, ... | | Authors: | Ehara, H, Kujirai, T, Shirouzu, M, Kurumizaka, H, Sekine, S. | | Deposit date: | 2022-05-15 | | Release date: | 2022-09-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science, 377, 2022
|
|
2H1E
 
 | |
6ZHY
 
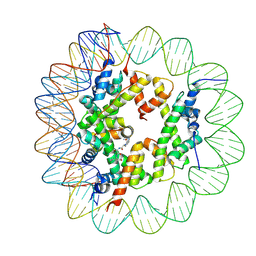 | | Cryo-EM structure of the regulatory linker of ALC1 bound to the nucleosome's acidic patch: hexasome class. | | Descriptor: | Chromodomain-helicase-DNA-binding protein 1-like, DNA (110-MER) Widom 601 sequence, Histone H2A type 1, ... | | Authors: | Bacic, L, Gaullier, G, Deindl, S. | | Deposit date: | 2020-06-24 | | Release date: | 2020-12-23 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch.
Cell Rep, 33, 2020
|
|
6Z6P
 
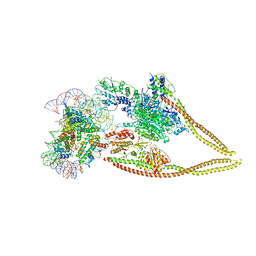 | | HDAC-PC-Nuc | | Descriptor: | DNA (145-MER), HDA1 complex subunit 2, HDA1 complex subunit 3,HDA1 complex subunit 3, ... | | Authors: | Lee, J.-H, Bollschweiler, D, Schaefer, T, Huber, R. | | Deposit date: | 2020-05-28 | | Release date: | 2021-02-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structural basis for the regulation of nucleosome recognition and HDAC activity by histone deacetylase assemblies.
Sci Adv, 7, 2021
|
|
7NKY
 
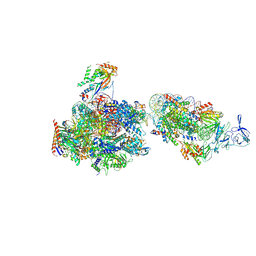 | | RNA Polymerase II-Spt4/5-nucleosome-FACT structure | | Descriptor: | Chromatin elongation factor SPT4, DNA (138-MER), DNA (148-MER), ... | | Authors: | Farnung, L, Ochmann, M, Engeholm, M, Cramer, P. | | Deposit date: | 2021-02-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of nucleosome transcription mediated by Chd1 and FACT.
Nat.Struct.Mol.Biol., 28, 2021
|
|
