4IUC
 
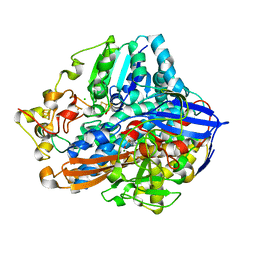 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its as-isolated form - oxidized state 2 | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Frielingsdorf, S, Schmidt, A, Fritsch, J, Lenz, O, Scheerer, P. | | Deposit date: | 2013-01-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reversible [4Fe-3S] cluster morphing in an O2-tolerant [NiFe] hydrogenase.
Nat.Chem.Biol., 10, 2014
|
|
4CI0
 
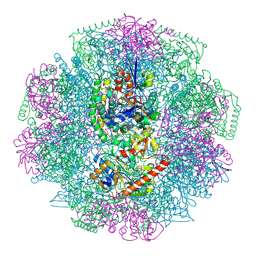 | | Electron cryo-microscopy of F420-reducing NiFe hydrogenase Frh | | Descriptor: | F420-REDUCING HYDROGENASE, SUBUNIT ALPHA, SUBUNIT BETA, ... | | Authors: | Allegretti, M, Mills, D.J, McMullan, G, Kuehlbrandt, W, Vonck, J. | | Deposit date: | 2013-12-05 | | Release date: | 2014-02-26 | | Last modified: | 2019-11-20 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Atomic Model of the F420-Reducing [Nife] Hydrogenase by Electron Cryo-Electron Microscopy Using a Direct Electron Detector.
Elife, 3, 2014
|
|
4C3O
 
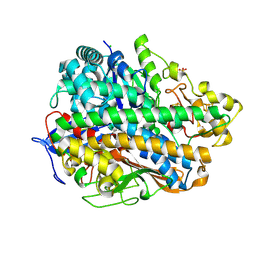 | | Structure and function of an oxygen tolerant NiFe hydrogenase from Salmonella | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Bowman, L, Flanagan, L, Fyfe, P.K, Parkin, A, Hunter, W.N, Sargent, F. | | Deposit date: | 2013-08-26 | | Release date: | 2014-01-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | How the Structure of the Large Subunit Controls Function in an Oxygen-Tolerant [Nife]-Hydrogenase.
Biochem.J., 458, 2014
|
|
4KO3
 
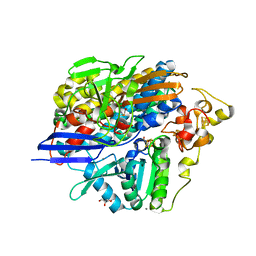 | | Low X-ray dose structure of anaerobically purified Dm. baculatum [NiFeSe]-hydrogenase after crystallization under air | | Descriptor: | CALCIUM ION, CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, ... | | Authors: | Volbeda, A, Cavazza, C, Fontecilla-Camps, J.C. | | Deposit date: | 2013-05-11 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural foundations for the O2 resistance of Desulfomicrobium baculatum [NiFeSe]-hydrogenase.
Chem.Commun.(Camb.), 49, 2013
|
|
4KN9
 
 | | High-resolution structure of H2-activated anaerobically purified Dm. baculatum [NiFeSe]-hydrogenase after crystallization under air | | Descriptor: | CALCIUM ION, CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, ... | | Authors: | Volbeda, A, Cavazza, C, Fontecilla-Camps, J.C. | | Deposit date: | 2013-05-09 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural foundations for the O2 resistance of Desulfomicrobium baculatum [NiFeSe]-hydrogenase.
Chem.Commun.(Camb.), 49, 2013
|
|
4KO2
 
 | | Low X-ray dose structure of H2-activated anaerobically purified Dm. baculatum [NiFeSe]-hydrogenase after crystallization under air | | Descriptor: | CALCIUM ION, CARBONMONOXIDE-(DICYANO) IRON, GLYCEROL, ... | | Authors: | Volbeda, A, Cavazza, C, Fontecilla-Camps, J.C. | | Deposit date: | 2013-05-11 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural foundations for the O2 resistance of Desulfomicrobium baculatum [NiFeSe]-hydrogenase.
Chem. Commun. (Camb.), 49, 2013
|
|
4KO4
 
 | | High X-ray dose structure of anaerobically purified Dm. baculatum [NiFeSe]-hydrogenase after crystallization under air | | Descriptor: | CALCIUM ION, CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, ... | | Authors: | Volbeda, A, Cavazza, C, Fontecilla-Camps, J.C. | | Deposit date: | 2013-05-11 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural foundations for the O2 resistance of Desulfomicrobium baculatum [NiFeSe]-hydrogenase.
Chem.Commun.(Camb.), 49, 2013
|
|
4KL8
 
 | | High-resolution structure of anaerobically purified Dm. baculatum [NiFeSe]-hydrogenase after crystallization under air | | Descriptor: | CALCIUM ION, CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, ... | | Authors: | Volbeda, A, Cavazza, C, Fontecilla-Camps, J.C. | | Deposit date: | 2013-05-07 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural foundations for the O2 resistance of Desulfomicrobium baculatum [NiFeSe]-hydrogenase.
Chem.Commun.(Camb.), 49, 2013
|
|
4KO1
 
 | | High X-ray dose structure of H2-activated anaerobically purified Dm. baculatum [NiFeSe]-hydrogenase after crystallization under air | | Descriptor: | CALCIUM ION, CARBONMONOXIDE-(DICYANO) IRON, GLYCEROL, ... | | Authors: | Volbeda, A, Cavazza, C, Fontecilla-Camps, J.C. | | Deposit date: | 2013-05-11 | | Release date: | 2013-07-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural foundations for the O2 resistance of Desulfomicrobium baculatum [NiFeSe]-hydrogenase.
Chem.Commun.(Camb.), 49, 2013
|
|
3ZE7
 
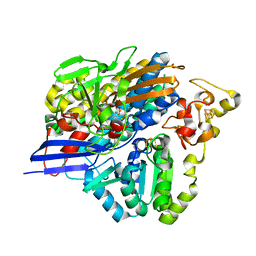 | | 3D structure of the Ni-Fe-Se hydrogenase from D. vulgaris Hildenborough in the reduced state at 1.95 Angstroms | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, HYDROSULFURIC ACID, ... | | Authors: | Marques, M.C, Coelho, R, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Redox State-Dependent Changes in the Crystal Structure of [Nifese] Hydrogenase from Desulfovibrio Vulgaris Hildenborough
Int.J.Hydrogen Energy, 38, 2013
|
|
3ZE9
 
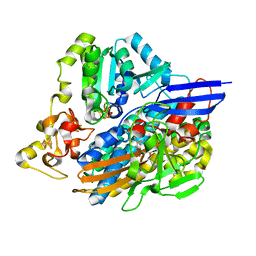 | | 3D structure of the NiFeSe hydrogenase from D. vulgaris Hildenborough in the oxidized as-isolated state at 1.33 Angstroms | | Descriptor: | BIS-(MU-2-OXO),[(MU-3--SULFIDO)-BIS(MU-2--SULFIDO)-TRIS(CYS-S)-TRI-IRON] (AQUA)(GLU-O)IRON(II), CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, ... | | Authors: | Marques, M.C, Coelho, R, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Redox State-Dependent Changes in the Crystal Structure of [Nifese] Hydrogenase from Desulfovibrio Vulgaris Hildenborough
Int.J.Hydrogen Energy, 38, 2013
|
|
3ZEA
 
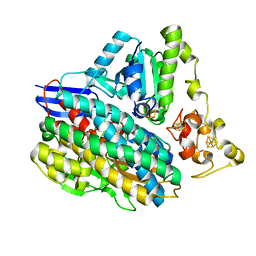 | | 3D structure of the NiFeSe hydrogenase from D. vulgaris Hildenborough in the reduced state at 1.82 Angstroms | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE (II) ION, HYDROSULFURIC ACID, ... | | Authors: | Marques, M.C, Coelho, R, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Redox State-Dependent Changes in the Crystal Structure of [Nifese] Hydrogenase from Desulfovibrio Vulgaris Hildenborough
Int.J.Hydrogen Energy, 38, 2013
|
|
3ZE8
 
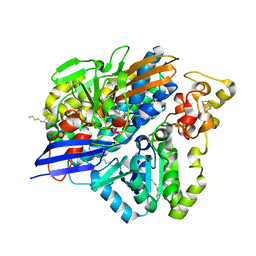 | | 3D structure of the Ni-Fe-Se hydrogenase from D. vulgaris Hildenborough in the reduced state at 1.95 Angstroms | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE (II) ION, ... | | Authors: | Marques, M.C, Coelho, R, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Redox State-Dependent Changes in the Crystal Structure of [Nifese] Hydrogenase from Desulfovibrio Vulgaris Hildenborough
Int.J.Hydrogen Energy, 38, 2013
|
|
3ZE6
 
 | | 3D structure of the Ni-Fe-Se hydrogenase from D. vulgaris Hildenborough in the as-isolated oxidized state at 1.50 Angstroms | | Descriptor: | 3-[DODECYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, BIS-(MU-2-OXO),[(MU-3--SULFIDO)-BIS(MU-2--SULFIDO)-TRIS(CYS-S)-TRI-IRON] (AQUA)(GLU-O)IRON(II), CARBONMONOXIDE-(DICYANO) IRON, ... | | Authors: | Marques, M.C, Coelho, R, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2012-12-03 | | Release date: | 2013-06-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox State-Dependent Changes in the Crystal Structure of [Nifese] Hydrogenase from Desulfovibrio Vulgaris Hildenborough
Int.J.Hydrogen Energy, 2013
|
|
3ZFS
 
 | | Cryo-EM structure of the F420-reducing NiFe-hydrogenase from a methanogenic archaeon with bound substrate | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, COENZYME F420, F420-REDUCING HYDROGENASE, ... | | Authors: | Mills, D.J, Vitt, S, Strauss, M, Shima, S, Vonck, J. | | Deposit date: | 2012-12-12 | | Release date: | 2013-03-06 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | De Novo Modeling of the F420-Reducing [Nife]-Hydrogenase from a Methanogenic Archaeon by Cryo-Electron Microscopy
Elife, 2, 2013
|
|
4GD3
 
 | | Structure of E. coli hydrogenase-1 in complex with cytochrome b | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2012-07-31 | | Release date: | 2013-01-02 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of the O(2)-Tolerant Membrane-Bound Hydrogenase 1 from Escherichia coli in Complex with Its Cognate Cytochrome b.
Structure, 21, 2013
|
|
3USC
 
 | | Crystal Structure of E. coli hydrogenase-1 in a ferricyanide-oxidized form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2011-11-23 | | Release date: | 2012-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UQY
 
 | | H2-reduced structure of E. coli hydrogenase-1 | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2011-11-21 | | Release date: | 2012-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3USE
 
 | | Crystal Structure of E. coli hydrogenase-1 in its as-isolated form | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Volbeda, A, Fontecilla-Camps, J.C, Darnault, C. | | Deposit date: | 2011-11-23 | | Release date: | 2012-03-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | X-ray crystallographic and computational studies of the O2-tolerant [NiFe]-hydrogenase 1 from Escherichia coli.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3RGW
 
 | | Crystal structure at 1.5 A resolution of an H2-reduced, O2-tolerant hydrogenase from Ralstonia eutropha unmasks a novel iron-sulfur cluster | | Descriptor: | FE3-S4 CLUSTER, FE4-S3 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Scheerer, P, Fritsch, J, Frielingsdorf, S, Kroschinsky, S, Friedrich, B, Lenz, O, Spahn, C.M.T. | | Deposit date: | 2011-04-10 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of an oxygen-tolerant hydrogenase uncovers a novel iron-sulphur centre.
Nature, 479, 2011
|
|
3AYX
 
 | | Membrane-bound respiratory [NiFe] hydrogenase from Hydrogenovibrio marinus in an H2-reduced condition | | Descriptor: | CARBON MONOXIDE, CYANIDE ION, FE (II) ION, ... | | Authors: | Shomura, Y, Yoon, K.S, Nishihara, H, Higuchi, Y. | | Deposit date: | 2011-05-20 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase
Nature, 479, 2011
|
|
3AYZ
 
 | | Membrane-bound respiratory [NiFe] hydrogenase from Hydrogenovibrio marinus in an air-oxidized condition | | Descriptor: | CARBON MONOXIDE, CYANIDE ION, FE (II) ION, ... | | Authors: | Shomura, Y, Yoon, K.S, Nishihara, H, Higuchi, Y. | | Deposit date: | 2011-05-20 | | Release date: | 2011-10-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase
Nature, 479, 2011
|
|
3MYR
 
 | | Crystal structure of [NiFe] hydrogenase from Allochromatium vinosum in its Ni-A state | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, Hydrogenase (NiFe) small subunit HydA, ... | | Authors: | Ogata, H, Kellers, P, Lubitz, W. | | Deposit date: | 2010-05-11 | | Release date: | 2010-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of the [NiFe] hydrogenase from the photosynthetic bacterium Allochromatium vinosum: characterization of the oxidized enzyme (Ni-A state).
J.Mol.Biol., 402, 2010
|
|
2WPN
 
 | | Structure of the oxidised, as-isolated NiFeSe hydrogenase from D. vulgaris Hildenborough | | Descriptor: | 3-[DODECYL(DIMETHYL)AMMONIO]PROPANE-1-SULFONATE, BIS-(MU-2-OXO),[(MU-3--SULFIDO)-BIS(MU-2--SULFIDO)-TRIS(CYS-S)-TRI-IRON] (AQUA)(GLU-O)IRON(II), CARBONMONOXIDE-(DICYANO) IRON, ... | | Authors: | Marques, M.C, Coelho, R, De Lacey, A.L, Pereira, I.A.C, Matias, P.M. | | Deposit date: | 2009-08-07 | | Release date: | 2010-01-12 | | Last modified: | 2019-07-10 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | The three-dimensional structure of [NiFeSe] hydrogenase from Desulfovibrio vulgaris Hildenborough: a hydrogenase without a bridging ligand in the active site in its oxidised, "as-isolated" state.
J.Mol.Biol., 396, 2010
|
|
3H3X
 
 | | Structure of the V74M large subunit mutant of NI-FE hydrogenase in an oxidized state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martinez, N, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2009-04-17 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Introduction of methionines in the gas channel makes [NiFe] hydrogenase aero-tolerant
J.Am.Chem.Soc., 131, 2009
|
|
