1DLM
 
 | | STRUCTURE OF CATECHOL 1,2-DIOXYGENASE FROM ACINETOBACTER CALCOACETICUS NATIVE DATA | | Descriptor: | CATECHOL 1,2-DIOXYGENASE, FE (III) ION, [1-PENTADECANOYL-2-DECANOYL-GLYCEROL-3-YL]PHOSPHONYL CHOLINE | | Authors: | Vetting, M.W, Ohlendorf, D.H. | | Deposit date: | 1999-12-11 | | Release date: | 2000-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The 1.8 A crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker.
Structure Fold.Des., 8, 2000
|
|
1ZO9
 
 | | Crystal Structure Of The Wild Type Heme Domain Of P450BM-3 with N-palmitoylmethionine | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bifunctional P-450:NADPH-P450 reductase, GLYCEROL, ... | | Authors: | Hegda, A, Chen, B, Tomchick, D.R, Bondlela, M, Haines, D.C, Schaffer, N, Machius, M, Graham, S.E, Peterson, J.A. | | Deposit date: | 2005-05-12 | | Release date: | 2006-08-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Interactions of substrates at the surface of P450s can greatly enhance substrate potency.
Biochemistry, 46, 2007
|
|
1ZP5
 
 | | Crystal structure of the complex between MMP-8 and a N-hydroxyurea inhibitor | | Descriptor: | CALCIUM ION, N-{2-[(4'-CYANO-1,1'-BIPHENYL-4-YL)OXY]ETHYL}-N'-HYDROXY-N-METHYLUREA, Neutrophil collagenase, ... | | Authors: | Campestre, C, Agamennone, M, Tortorella, P, Preziuso, S, Biasone, A, Gavuzzo, E, Pochetti, G, Mazza, F, Tschesche, H, Gallina, C. | | Deposit date: | 2005-05-16 | | Release date: | 2005-12-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | N-Hydroxyurea as zinc binding group in matrix metalloproteinase inhibition: Mode of binding in a complex with MMP-8.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1LOJ
 
 | | Crystal structure of a Methanobacterial Sm-like archaeal protein (SmAP1) bound to uridine-5'-monophosphate (UMP) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, URIDINE, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Mura, C, Kozhukhovsky, A, Eisenberg, D. | | Deposit date: | 2002-05-06 | | Release date: | 2003-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The oligomerization and ligand-binding properties of Sm-like archaeal proteins (SmAPs)
Protein Sci., 12, 2003
|
|
1Z4R
 
 | | Human GCN5 Acetyltransferase | | Descriptor: | ACETYL COENZYME *A, General control of amino acid synthesis protein 5-like 2 | | Authors: | Dong, A, Bernstein, G, Schuetz, A, Antoshenko, T, Wu, H, Loppnau, P, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Plotnikov, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-16 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Crystal structure of a binary complex between human GCN5 histone acetyltransferase domain and acetyl coenzyme A
Proteins, 68, 2007
|
|
1M5H
 
 | | Formylmethanofuran:tetrahydromethanopterin formyltransferase from Archaeoglobus fulgidus | | Descriptor: | Formylmethanofuran--tetrahydromethanopterin formyltransferase, POTASSIUM ION | | Authors: | Mamat, B, Roth, A, Grimm, C, Ermler, U, Tziatzios, C, Schubert, D, Thauer, R.K, Shima, S. | | Deposit date: | 2002-07-09 | | Release date: | 2002-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures and enzymatic properties of three formyltransferases from archaea: environmental adaptation and evolutionary relationship.
Protein Sci., 11, 2002
|
|
1H8E
 
 | | (ADP.AlF4)2(ADP.SO4) bovine F1-ATPase (all three catalytic sites occupied) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BOVINE MITOCHONDRIAL F1-ATPASE, GLYCEROL, ... | | Authors: | Menz, R.I, Walker, J.E, Leslie, A.G.W. | | Deposit date: | 2001-02-02 | | Release date: | 2001-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Bovine Mitochondrial F1-ATPase with Nucleotide Bound to All Three Catalytic Sites: Implications for the Mechanism of Rotary Catalysis
Cell(Cambridge,Mass.), 106, 2001
|
|
1YP7
 
 | | Van der Waals Interactions Dominate Hydrophobic Association in a Protein Binding Site Occluded From Solvent Water | | Descriptor: | CADMIUM ION, MAJOR URINARY PROTEIN 1 | | Authors: | Barratt, E, Bingham, R.J, Warner, D.J, Laughton, C.A, Phillips, S.E.V, Homans, S.W. | | Deposit date: | 2005-01-30 | | Release date: | 2005-08-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Van der Waals Interactions Dominate Ligand-Protein Association in a Protein Binding Site Occluded from Solvent Water
J.Am.Chem.Soc., 127, 2005
|
|
1DRW
 
 | | ESCHERICHIA COLI DHPR/NHDH COMPLEX | | Descriptor: | DIHYDRODIPICOLINATE REDUCTASE, NICOTINAMIDE PURIN-6-OL-DINUCLEOTIDE | | Authors: | Reddy, S.G, Scapin, G, Blanchard, J.S. | | Deposit date: | 1996-06-28 | | Release date: | 1997-01-27 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interaction of pyridine nucleotide substrates with Escherichia coli dihydrodipicolinate reductase: thermodynamic and structural analysis of binary complexes.
Biochemistry, 35, 1996
|
|
2J5B
 
 | | Structure of the Tyrosyl tRNA synthetase from Acanthamoeba polyphaga Mimivirus complexed with tyrosynol | | Descriptor: | 4-[(2S)-2-amino-3-hydroxypropyl]phenol, TYROSYL-TRNA SYNTHETASE | | Authors: | Abergel, C, Rudinger-thirion, J, Giege, R, Claverie, J.M. | | Deposit date: | 2006-09-13 | | Release date: | 2007-09-25 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Virus-Encoded Aminoacyl-tRNA Synthetases: Structural and Functional Characterization of Mimivirus Tyrrs and Metrs.
J.Virol., 81, 2007
|
|
1L7Y
 
 | | Solution NMR Structure of C. elegans Protein ZK652.3. NORTHEAST STRUCTURAL GENOMICS CONSORTIUM TARGET WR41. | | Descriptor: | HYPOTHETICAL PROTEIN ZK652.3 | | Authors: | Cort, J.R, Chiang, Y, Zheng, D, Montelione, G.T, Kennedy, M.A, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2002-03-18 | | Release date: | 2002-08-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of conserved eukaryotic protein ZK652.3 from C. elegans: a ubiquitin-like fold.
Proteins, 48, 2002
|
|
2A78
 
 | | Crystal structure of the C3bot-RalA complex reveals a novel type of action of a bacterial exoenzyme | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Mono-ADP-ribosyltransferase C3, ... | | Authors: | Pautsch, A, Vogelsgesang, M, Trankle, J, Herrmann, C, Aktories, K. | | Deposit date: | 2005-07-05 | | Release date: | 2005-10-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structure of the C3bot-RalA complex reveals a novel type of action of a bacterial exoenzyme.
Embo J., 24, 2005
|
|
1YP6
 
 | | Van der Waals Interactions Dominate Hydrophobic Association in a Protein Binding Site Occluded From Solvent Water | | Descriptor: | 2-ISOBUTYL-3-METHOXYPYRAZINE, CADMIUM ION, CHLORIDE ION, ... | | Authors: | Barratt, E, Bingham, R.J, Warner, D.J, Laughton, C.A, Phillips, S.E.V, Homans, S.W. | | Deposit date: | 2005-01-30 | | Release date: | 2005-08-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Van der Waals Interactions Dominate Ligand-Protein Association in a Protein Binding Site Occluded from Solvent Water
J.Am.Chem.Soc., 127, 2005
|
|
1ZGR
 
 | |
1ZGS
 
 | |
2FSP
 
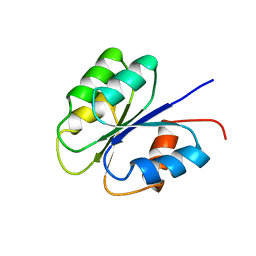 | | NMR SOLUTION STRUCTURE OF BACILLUS SUBTILIS SPO0F PROTEIN, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | STAGE 0 SPORULATION PROTEIN F | | Authors: | Feher, V.A, Skelton, N.J, Dahlquist, F.W, Cavanagh, J. | | Deposit date: | 1997-06-06 | | Release date: | 1997-12-10 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | High-resolution NMR structure and backbone dynamics of the Bacillus subtilis response regulator, Spo0F: implications for phosphorylation and molecular recognition.
Biochemistry, 36, 1997
|
|
1LPV
 
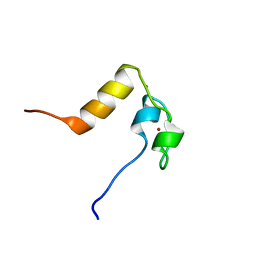 | | DROSOPHILA MELANOGASTER DOUBLESEX (DSX), NMR, 18 STRUCTURES | | Descriptor: | Doublesex protein, ZINC ION | | Authors: | Zhu, L, Wilken, J, Phillips, N, Narendra, U, Chan, G, Stratton, S, Kent, S, Weiss, M.A. | | Deposit date: | 2002-05-08 | | Release date: | 2002-10-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Sexual dimorphism in diverse metazoans is regulated by a novel class of intertwined zinc fingers.
Genes Dev., 14, 2000
|
|
1CQT
 
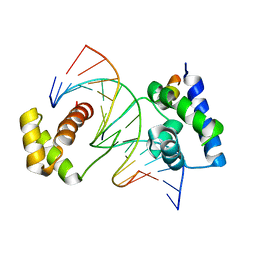 | | CRYSTAL STRUCTURE OF A TERNARY COMPLEX CONTAINING AN OCA-B PEPTIDE, THE OCT-1 POU DOMAIN, AND AN OCTAMER ELEMENT | | Descriptor: | DNA (5'-D(*AP*CP*CP*TP*TP*AP*TP*TP*TP*GP*CP*AP*TP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*AP*TP*GP*CP*AP*AP*AP*TP*AP*AP*GP*G)-3'), POU DOMAIN, ... | | Authors: | Chasman, D.I, Cepek, K, Sharp, P.A, Pabo, C.O. | | Deposit date: | 1999-08-11 | | Release date: | 1999-11-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of an OCA-B peptide bound to an Oct-1 POU domain/octamer DNA complex: specific recognition of a protein-DNA interface.
Genes Dev., 13, 1999
|
|
2J9I
 
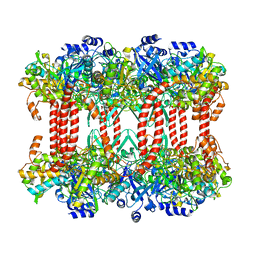 | | Lengsin is a survivor of an ancient family of class I glutamine synthetases in eukaryotes that has undergone evolutionary re- engineering for a tissue-specific role in the vertebrate eye lens. | | Descriptor: | GLUTAMATE-AMMONIA LIGASE DOMAIN-CONTAINING PROTEIN 1 | | Authors: | Wyatt, K, White, H.E, Wang, L, Bateman, O.A, Slingsby, C, Orlova, E.V, Wistow, G. | | Deposit date: | 2006-11-09 | | Release date: | 2006-12-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Lengsin is a Survivor of an Ancient Family of Class I Glutamine Synthetases Re-Engineered by Evolution for a Role in the Vertebrate Lens.
Structure, 14, 2006
|
|
1DRV
 
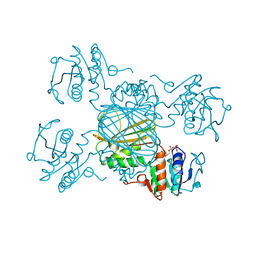 | | ESCHERICHIA COLI DHPR/ACNADH COMPLEX | | Descriptor: | 3-ACETYLPYRIDINE ADENINE DINUCLEOTIDE, DIHYDRODIPICOLINATE REDUCTASE | | Authors: | Reddy, S.G, Scapin, G, Blanchard, J.S. | | Deposit date: | 1996-06-28 | | Release date: | 1997-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interaction of pyridine nucleotide substrates with Escherichia coli dihydrodipicolinate reductase: thermodynamic and structural analysis of binary complexes.
Biochemistry, 35, 1996
|
|
1Z00
 
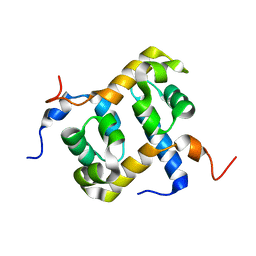 | | Solution structure of the C-terminal domain of ERCC1 complexed with the C-terminal domain of XPF | | Descriptor: | DNA excision repair protein ERCC-1, DNA repair endonuclease XPF | | Authors: | Tripsianes, K, Folkers, G, Ab, E, Das, D, Odijk, H, Jaspers, N.G.J, Hoeijmakers, J.H.J, Kaptein, R, Boelens, R. | | Deposit date: | 2005-03-01 | | Release date: | 2005-12-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Human ERCC1/XPF Interaction Domains Reveals a Complementary Role for the Two Proteins in Nucleotide Excision Repair
Structure, 13, 2005
|
|
1OCN
 
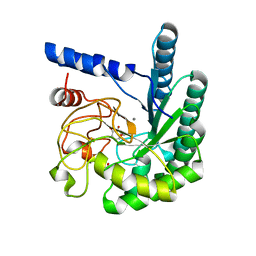 | | Mutant D416A of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with a cellobio-derived isofagomine at 1.3 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, CALCIUM ION, ... | | Authors: | Varrot, A, Macdonald, J, Stick, R.V, Pell, G, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2003-02-09 | | Release date: | 2003-05-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Distortion of a Cellobio-Derived Isofagomine Highlights the Potential Conformational Itinerary of Inverting Beta-Glucosidases
Chem.Commun.(Camb.), 21, 2003
|
|
1NW1
 
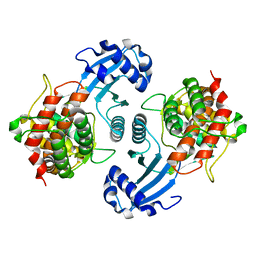 | | Crystal Structure of Choline Kinase | | Descriptor: | CALCIUM ION, Choline kinase (49.2 kD) | | Authors: | Peisach, D, Gee, P, Kent, C, Xu, Z. | | Deposit date: | 2003-02-05 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The Crystal Structure of Choline Kinase Reveals a Eukaryotic Protein Kinase Fold
Structure, 11, 2003
|
|
1OYS
 
 | |
5TLQ
 
 | | Model structure of the oxidized PaDsbA1 and 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine complex | | Descriptor: | 3-[(2-methylbenzyl)sulfanyl]-4H-1,2,4-triazol-4-amine, Thiol:disulfide interchange protein DsbA | | Authors: | Mohanty, B, Rimmer, K.A, McMahon, R.M, Headey, S.J, Vazirani, M, Shouldice, S.R, Coincon, M, Tay, S, Morton, C.J, Simpson, J.S, Martin, J.L, Scanlon, M.S. | | Deposit date: | 2016-10-11 | | Release date: | 2017-04-12 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | Fragment library screening identifies hits that bind to the non-catalytic surface of Pseudomonas aeruginosa DsbA1.
PLoS ONE, 12, 2017
|
|
