4TRJ
 
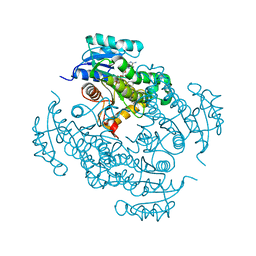 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (INHA) complexed with N-(3-bromophenyl)-1-cyclohexyl-5-oxopyrrolidine-3-carboxamide, refined with new ligand restraints | | Descriptor: | (3S)-N-(3-BROMOPHENYL)-1-CYCLOHEXYL-5-OXOPYRROLIDINE-3-CARBOXAMIDE, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | He, X, Alian, A, Stroud, R.M, Ortiz de Montellano, P.R. | | Deposit date: | 2014-06-17 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Pyrrolidine carboxamides as a novel class of inhibitors of enoyl acyl carrier protein reductase from Mycobacterium tuberculosis
J. Med. Chem., 2006
|
|
6DVI
 
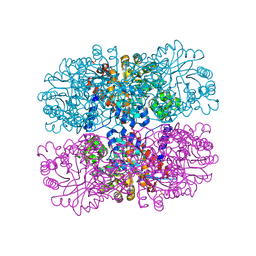 | |
4G3U
 
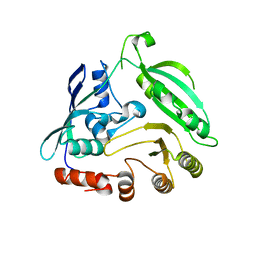 | |
1FZT
 
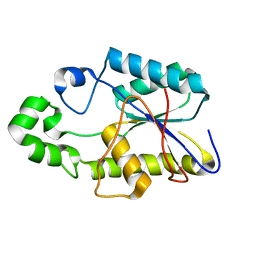 | | SOLUTION STRUCTURE AND DYNAMICS OF AN OPEN B-SHEET, GLYCOLYTIC ENZYME-MONOMERIC 23.7 KDA PHOSPHOGLYCERATE MUTASE FROM SCHIZOSACCHAROMYCES POMBE | | Descriptor: | PHOSPHOGLYCERATE MUTASE | | Authors: | Uhrinova, S, Uhrin, D, Nairn, J, Price, N.C, Fothergill-Gilmore, L.A. | | Deposit date: | 2000-10-04 | | Release date: | 2001-03-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of an open beta-sheet, glycolytic enzyme, monomeric 23.7 kDa phosphoglycerate mutase from Schizosaccharomyces pombe.
J.Mol.Biol., 306, 2001
|
|
4GO5
 
 | |
4GO7
 
 | |
6AC6
 
 | | Ab initio crystal structure of Selenomethionine labelled Mycobacterium smegmatis Mfd | | Descriptor: | Mycobacterium smegmatis Mfd, SULFATE ION | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.989 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
4RAE
 
 | |
6AC8
 
 | | Crystal structure of Mycobacterium smegmatis Mfd at 2.75 A resolution | | Descriptor: | Mycobacterium smegmatis Mfd, SULFATE ION | | Authors: | Putta, S, Fox, G.C, Walsh, M.A, Rao, D.N, Nagaraja, V, Natesh, R. | | Deposit date: | 2018-07-25 | | Release date: | 2019-08-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for nucleotide-mediated remodelling mechanism of Mycobacterium Mfd
To Be Published
|
|
4R2N
 
 | | Crystal structure of Rv3772 in complex with its substrate | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, PHENYLALANINE, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nasir, N, Anant, A, Vyas, R, Biswal, B.K. | | Deposit date: | 2014-08-12 | | Release date: | 2015-08-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis HspAT and ArAT reveal structural basis of their distinct substrate specificities
Sci Rep, 6, 2016
|
|
4R8D
 
 | | Crystal structure of Rv1600 encoded aminotransferase in complex with PLP-MES from Mycobacterium tuberculosis | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Histidinol-phosphate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Nasir, N, Anant, A, Vyas, R, Biswal, B.K. | | Deposit date: | 2014-09-01 | | Release date: | 2015-09-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Rv1600 encoded aminotransferase in complex with PLP-MES from Mycobacterium tuberculosis
To be Published
|
|
5DB4
 
 | |
2WU8
 
 | |
2YGA
 
 | | E88G-N92L Mutant of N-Term HSP90 complexed with Geldanamycin | | Descriptor: | ATP-DEPENDENT MOLECULAR CHAPERONE HSP82, GELDANAMYCIN | | Authors: | Roe, S.M, Prodromou, C, Pearl, L.H. | | Deposit date: | 2011-04-12 | | Release date: | 2011-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Features of the Streptomyces Hygroscopicus Htpg Reveal How Partial Geldanamycin Resistance Can Arise with Mutation to the ATP Binding Pocket of a Eukaryotic Hsp90.
Faseb J., 25, 2011
|
|
4R5Z
 
 | | Crystal structure of Rv3772 encoded aminotransferase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Nasir, N, Anant, A, Vyas, R, Biswal, B.K. | | Deposit date: | 2014-08-22 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis HspAT and ArAT reveal structural basis of their distinct substrate specificities
Sci Rep, 6, 2016
|
|
6D8I
 
 | |
6D8J
 
 | |
3QPL
 
 | | G106W mutant of EthR from Mycobacterium tuberculosis | | Descriptor: | HTH-type transcriptional regulator EthR | | Authors: | Carette, X, Willery, E, Hoos, S, Lecat-Guillet, N, Frenois, F, Dirie, B, Villeret, V, England, P, Deprez, B, Locht, C, Willand, N, Baulard, A. | | Deposit date: | 2011-02-14 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural activation of the transcriptional repressor EthR from Mycobacterium tuberculosis by single amino acid change mimicking natural and synthetic ligands.
Nucleic Acids Res., 40, 2012
|
|
8D6Y
 
 | |
8D6X
 
 | |
4QVB
 
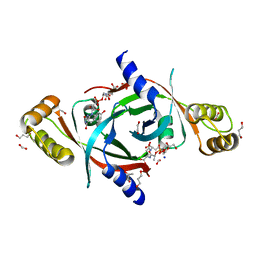 | | Mycobacterium tuberculosis protein Rv1155 in complex with co-enzyme F420 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, COENZYME F420, ... | | Authors: | Mashalidis, E.H, Gittis, A.G, Tomczak, A, Abell, C, Barry III, C.E, Garboczi, D.N. | | Deposit date: | 2014-07-14 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Molecular insights into the binding of coenzyme F420 to the conserved protein Rv1155 from Mycobacterium tuberculosis.
Protein Sci., 24, 2015
|
|
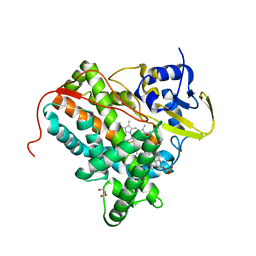
4TRI
 
 | |
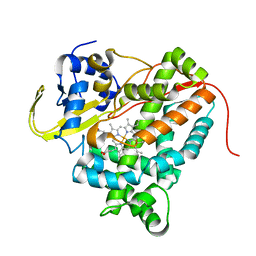
4UAX
 
 | |
1P0H
 
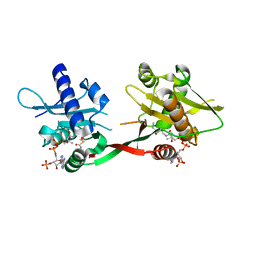 | | Crystal Structure of Rv0819 from Mycobacterium Tuberculosis MshD-Mycothiol Synthase Coenzyme A Complex | | Descriptor: | ACETYL COENZYME *A, COENZYME A, hypothetical protein Rv0819 | | Authors: | Vetting, M.W, Roderick, S.L, Yu, M, Blanchard, J.S. | | Deposit date: | 2003-04-10 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of mycothiol synthase (Rv0819) from Mycobacterium tuberculosis shows structural homology to the GNAT family of N-acetyltransferases.
Protein Sci., 12, 2003
|
|
1OZP
 
 | | Crystal Structure of Rv0819 from Mycobacterium tuberculosis MshD-Mycothiol Synthase Acetyl-Coenzyme A Complex. | | Descriptor: | ACETYL COENZYME *A, hypothetical protein Rv0819 | | Authors: | Vetting, M.W, Roderick, S.L, Yu, M, Blanchard, J.S. | | Deposit date: | 2003-04-09 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of mycothiol synthase (Rv0819) from Mycobacterium tuberculosis shows structural homology to the GNAT family of N-acetyltransferases.
Protein Sci., 12, 2003
|
|
