8VAW
 
 | |
8REF
 
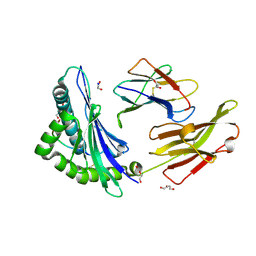 | | Crystal structure of HLA B*13:01 in complex with SVLNDILARL, an 10-mer epitope from SARS-CoV-2 Spike (S975-984) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-11 | | Release date: | 2024-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
8RE4
 
 | |
8RED
 
 | |
8REE
 
 | |
8REC
 
 | |
8REA
 
 | |
8REB
 
 | |
8VA2
 
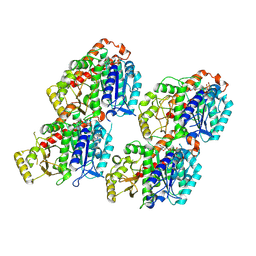 | |
8VA0
 
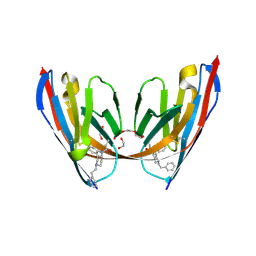 | | X-ray crystal structure of JGFN4 N76D complexed with fentanyl in dimer form | | Descriptor: | 1,2-ETHANEDIOL, JGFN4, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide, ... | | Authors: | Shi, K, Moller, N, Aihara, H. | | Deposit date: | 2023-12-10 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification and biophysical characterization of a novel domain-swapped camelid antibody specific for fentanyl.
J.Biol.Chem., 300, 2024
|
|
8V9X
 
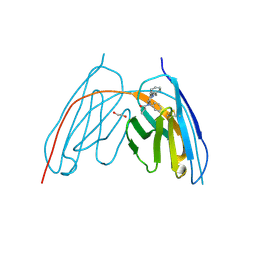 | | X-ray crystal structure of JGFN4 complex with fentanyl | | Descriptor: | 1,2-ETHANEDIOL, JGFN4, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide | | Authors: | Moller, N, Shi, K, Aihara, H. | | Deposit date: | 2023-12-10 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and biophysical characterization of a novel domain-swapped camelid antibody specific for fentanyl.
J.Biol.Chem., 300, 2024
|
|
8V9Y
 
 | |
8V9Z
 
 | |
8V9W
 
 | | X-ray crystal structure of JGFN4 complexed with fentanyl | | Descriptor: | 1,2-ETHANEDIOL, JGFN4, N-phenyl-N-[1-(2-phenylethyl)piperidin-4-yl]propanamide | | Authors: | Moller, N, Shi, K, Aihara, H. | | Deposit date: | 2023-12-10 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Identification and biophysical characterization of a novel domain-swapped camelid antibody specific for fentanyl.
J.Biol.Chem., 300, 2024
|
|
8RDZ
 
 | |
8RDF
 
 | |
8RDU
 
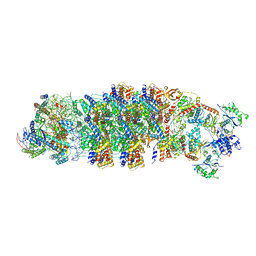 | | Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K composite map | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, LE, MAGNESIUM ION, ... | | Authors: | Tenjo-Castano, F, Mesa, P, Montoya, G. | | Deposit date: | 2023-12-08 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
8V9O
 
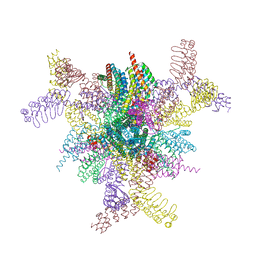 | | Imaging scaffold engineered to bind the therapeutic protein target BARD1 | | Descriptor: | CALCIUM ION, Tetrahedral Nanocage Cage Component Fused to Anti-BARD1 Darpin, Tetrahedral Nanocage Cage, ... | | Authors: | Agdanowski, M.P, Castells-Graells, R, Sawaya, M.R, Yeates, T.O, Arbing, M.A. | | Deposit date: | 2023-12-08 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | X-ray crystal structure of a designed rigidified imaging scaffold in the ligand-free conformation.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8RDV
 
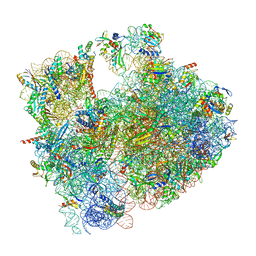 | | Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factor Balon, mRNA and P-site tRNA (structure 2). | | Descriptor: | 16S rRNA, 23S rRNA, 5S rRNA, ... | | Authors: | Helena-Bueno, K, Rybak, M.Y, Gagnon, M.G, Hill, C.H, Melnikov, S.V. | | Deposit date: | 2023-12-08 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | A new family of bacterial ribosome hibernation factors.
Nature, 626, 2024
|
|
8RDW
 
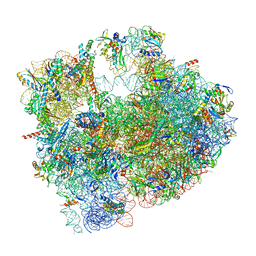 | | Cryo-EM structure of P. urativorans 70S ribosome in complex with hibernation factor Balon and EF-Tu(GDP) (structure 3). | | Descriptor: | 16S rRNA, 23S rRNA, 5S rRNA, ... | | Authors: | Helena-Bueno, K, Rybak, M.Y, Gagnon, M.G, Hill, C.H, Melnikov, S.V. | | Deposit date: | 2023-12-08 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | A new family of bacterial ribosome hibernation factors.
Nature, 626, 2024
|
|
8V9J
 
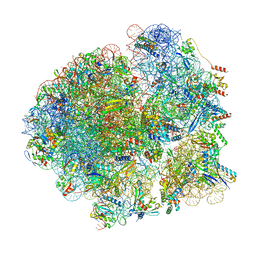 | | Cryo-EM structure of the Mycobacterium smegmatis 70S ribosome in complex with hibernation factor Msmeg1130 (Balon) (Structure 4) | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S14, ... | | Authors: | Rybak, M.Y, Helena-Bueno, K, Hill, C.H, Melnikov, S.V, Gagnon, M.G. | | Deposit date: | 2023-12-08 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | A new family of bacterial ribosome hibernation factors.
Nature, 626, 2024
|
|
8V9Q
 
 | | Crystal structure of mGalNAc-T1 in complex with the mucin glycopeptide Muc5AC-13, Mn2+, and UDP. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Samara, N.L, Collette, A.M. | | Deposit date: | 2023-12-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | An unusual dual sugar-binding lectin domain controls the substrate specificity of a mucin-type O-glycosyltransferase.
Sci Adv, 10, 2024
|
|
8V9M
 
 | | Human Ornithine Aminotransferase cocrystallized with its inhibitor, (R)-3-amino-5,5-difluorocyclohex-1-ene-1-carboxylic acid. | | Descriptor: | 3-fluoro-5-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]benzoic acid, GLYCEROL, Ornithine aminotransferase, ... | | Authors: | Vargas, A.L, Devitt, A, Kaley, N, Silverman, R, Liu, D. | | Deposit date: | 2023-12-08 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Design, Synthesis, and Mechanistic Studies of ( R )-3-Amino-5,5-difluorocyclohex-1-ene-1-carboxylic Acid as an Inactivator of Human Ornithine Aminotransferase.
Acs Chem.Biol., 19, 2024
|
|
8RDJ
 
 | | Plastid-encoded RNA polymerase transcription elongation complex (Integrated model) | | Descriptor: | DNA (81-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Webster, M.W, Pramanick, I, Vergara-Cruces, A. | | Deposit date: | 2023-12-08 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure of the plant plastid-encoded RNA polymerase.
Cell, 187, 2024
|
|
8RDD
 
 | |
