3W2G
 
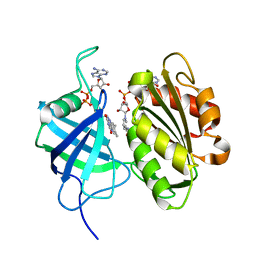 | | Crystal structure of fully reduced form of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3W5H
 
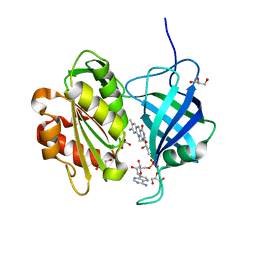 | | Ultra-high resolution structure of NADH-cytochrome b5 reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, NADH-cytochrome b5 reductase 3 | | Authors: | Takeda, K, Ohno, H, Kosugi, M, Takaba, K, Miki, K. | | Deposit date: | 2013-01-30 | | Release date: | 2013-07-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (0.78 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3W2I
 
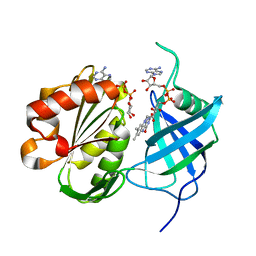 | | Crystal structure of re-oxidized form (60 min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3W2F
 
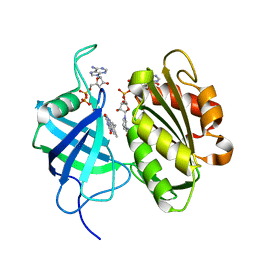 | | Crystal structure of oxidation intermediate (10 min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
3W5V
 
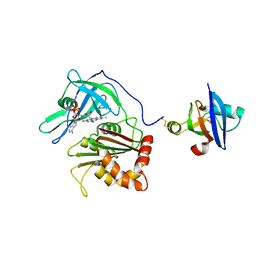 | | Cross-linked complex between Ferredoxin and Ferredoxin-NADP+ reductase | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin, ... | | Authors: | Kimata-Ariga, Y, Kubota-Kawai, H, Muraki, N, Hase, T, Kurisu, G. | | Deposit date: | 2013-02-06 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.81 Å) | | Cite: | Concentration-dependent oligomerization of cross-linked complexes between ferredoxin and ferredoxin-NADP(+) reductase
Biochem.Biophys.Res.Commun., 434, 2013
|
|
3W2H
 
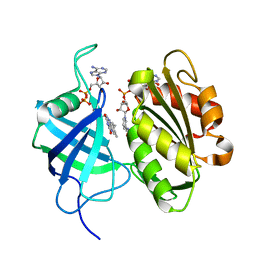 | | Crystal structure of oxidation intermediate (1min) of NADH-cytochrome b5 reductase from pig liver | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Yamada, M, Tamada, T, Matsumoto, F, Shoyama, Y, Kimura, S, Kuroki, R, Miki, K. | | Deposit date: | 2012-11-28 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Elucidations of the catalytic cycle of NADH-cytochrome b5 reductase by X-ray crystallography: new insights into regulation of efficient electron transfer
J.Mol.Biol., 425, 2013
|
|
8A1V
 
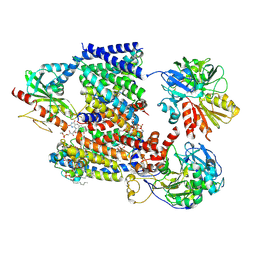 | | Sodium pumping NADH-quinone oxidoreductase with substrate Q2 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1W
 
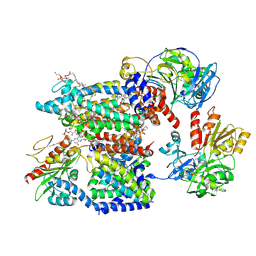 | | Sodium pumping NADH-quinone oxidoreductase with substrate Q1 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1T
 
 | | Sodium pumping NADH-quinone oxidoreductase | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1X
 
 | | Sodium pumping NADH-quinone oxidoreductase with inhibitor DQA | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1Y
 
 | | Sodium pumping NADH-quinone oxidoreductase with inhibitor HQNO | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8A1U
 
 | | Sodium pumping NADH-quinone oxidoreductase with substrates NADH and Q2 | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Hau, J.-L, Kaltwasser, S, Vonck, J, Fritz, G, Steuber, J. | | Deposit date: | 2022-06-02 | | Release date: | 2023-09-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AD3
 
 | | X-ray structure of NqrF(129-408)of Vibrio cholerae variant F406A | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, Na(+)-translocating NADH-quinone reductase subunit F | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8ACW
 
 | | X-ray structure of Na+-NQR from Vibrio cholerae at 3.4 A resolution | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AD4
 
 | | X-ray structure of NqrF(129-408)of Vibrio cholerae in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit F, ... | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8ACY
 
 | | X-ray structure of Na+-NQR from Vibrio cholerae at 3.5 A resolution | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AD5
 
 | | X-ray structure of NqrF(129-408)of Vibrio cholerae variant F406A | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, Na(+)-translocating NADH-quinone reductase subunit F, ... | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8AD0
 
 | |
