1AXO
 
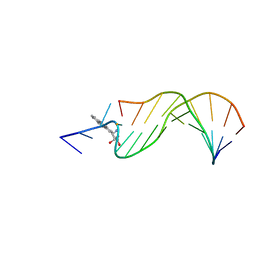 | | STRUCTURAL ALIGNMENT OF THE (+)-TRANS-ANTI-[BP]DG ADDUCT POSITIONED OPPOSITE DC AT A DNA TEMPLATE-PRIMER JUNCTION, NMR, 6 STRUCTURES | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, DNA DUPLEX D(AAC-[BP]G-CTACCATCC)D(GGATGGTAGC) | | Authors: | Feng, B, Gorin, A.A, Hingerty, B.E, Geacintov, N.E, Broyde, S, Patel, D.J. | | Deposit date: | 1997-10-16 | | Release date: | 1998-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural alignment of the (+)-trans-anti-benzo[a]pyrene-dG adduct positioned opposite dC at a DNA template-primer junction.
Biochemistry, 36, 1997
|
|
6AUU
 
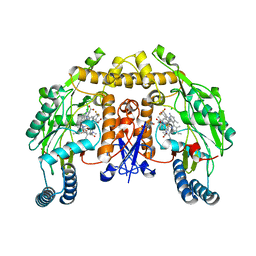 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(3-(3-(dimethylamino)propyl)-5-(trifluoromethyl)phenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{3-[3-(dimethylamino)propyl]-5-(trifluoromethyl)phenyl}ethyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Improvement of Cell Permeability of Human Neuronal Nitric Oxide Synthase Inhibitors Using Potent and Selective 2-Aminopyridine-Based Scaffolds with a Fluorobenzene Linker.
J. Med. Chem., 60, 2017
|
|
8A2K
 
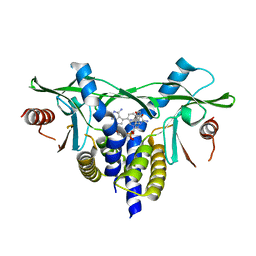 | | human STING in complex with 2'-3'-cyclic-GMP-7-deaza(4-[(2-naphthyloxy)methyl]phenyl)-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-[4-azanyl-5-[4-(naphthalen-1-yloxymethyl)phenyl]pyrrolo[2,3-d]pyrimidin-7-yl]-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-3~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Vavrina, Z, Brynda, J, Rezacova, P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
8A2H
 
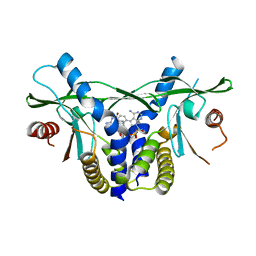 | | human STING in complex with 2',3'-cyclic-GMP-7-deazaphenyl-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-(4-azanyl-5-phenyl-pyrrolo[2,3-d]pyrimidin-7-yl)-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Smola, M, Vavrina, Z, Boura, E, Brynda, J. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
8A2J
 
 | | human STING in complex with 2'-3'-cyclic-GMP-7-deaza(4-biphenylyl)-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-[4-azanyl-5-(4-phenylphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Vavrina, Z, Brynda, J, Rezacova, P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
8A2I
 
 | | human STING in complex with 2'-3'-cyclic-GMP-7-deaza(4-(2-naphthyl)phenyl)-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-[4-azanyl-5-(4-naphthalen-2-ylphenyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-3~{H}-purin-6-one, Stimulator of interferon genes protein | | Authors: | Vavrina, Z, Brynda, J, Rezacova, P. | | Deposit date: | 2022-06-03 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Design, Synthesis, and Biochemical and Biological Evaluation of Novel 7-Deazapurine Cyclic Dinucleotide Analogues as STING Receptor Agonists.
J.Med.Chem., 65, 2022
|
|
6EV7
 
 | | Structure of E282D A. niger Fdc1 with prFMN in the iminium form | | Descriptor: | 1-deoxy-5-O-phosphono-1-(3,3,4,5-tetramethyl-9,11-dioxo-2,3,8,9,10,11-hexahydro-7H-quinolino[1,8-fg]pteridin-12-ium-7-y l)-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, Leys, D, Payne, K.A.P. | | Deposit date: | 2017-11-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | The role of conserved residues in Fdc decarboxylase in prenylated flavin mononucleotide oxidative maturation, cofactor isomerization, and catalysis.
J. Biol. Chem., 293, 2018
|
|
6EVA
 
 | | Structure of E277Q A. niger fdc1 in complex with a phenylpyruvate derived adduct to the prenylated flavin cofactor | | Descriptor: | 1-deoxy-5-O-phosphono-1-[(1S)-3,3,4,5-tetramethyl-9,11-dioxo-1-(phenylacetyl)-2,3,8,9,10,11-hexahydro-1H,7H-quinolino[1 ,8-fg]pteridin-7-yl]-D-ribitol, Ferulic acid decarboxylase 1, MANGANESE (II) ION, ... | | Authors: | Bailey, S.S, David, L, Payne, K.A.P. | | Deposit date: | 2017-11-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | The role of conserved residues in Fdc decarboxylase in prenylated flavin mononucleotide oxidative maturation, cofactor isomerization, and catalysis.
J. Biol. Chem., 293, 2018
|
|
5WM8
 
 | | Structure of the 10R (+)-cis-BP-dG modified Rev1 ternary complex | | Descriptor: | 1,2,3-TRIHYDROXY-1,2,3,4-TETRAHYDROBENZO[A]PYRENE, 1,2-ETHANEDIOL, 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Rechkoblit, O, Kolbanovsky, A, Landes, H, Geacintov, N.E, Aggarwal, A.K. | | Deposit date: | 2017-07-28 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Mechanism of error-free replication across benzo[a]pyrene stereoisomers by Rev1 DNA polymerase.
Nat Commun, 8, 2017
|
|
8DHD
 
 | | Neutron crystal structure of maltotetraose bound tmMBP | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein MalE2 | | Authors: | Cuneo, M.J, Shukla, S, Myles, D.A. | | Deposit date: | 2022-06-27 | | Release date: | 2022-10-12 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Mapping periplasmic binding protein oligosaccharide recognition with neutron crystallography.
Sci Rep, 12, 2022
|
|
7ZVR
 
 | | Crystal structure of the carotenoid-binding protein domain from silkworm Bombyx mori (BmCBP) complexed with zeaxanthin | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, Carotenoid-binding protein | | Authors: | Sluchanko, N.N, Boyko, K.M, Varfolomeeva, L.A, Slonimskiy, Y.B, Egorkin, N.A, Maksimov, E.G, Popov, V.O. | | Deposit date: | 2022-05-17 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the carotenoid binding and transport function of a START domain.
Structure, 30, 2022
|
|
1BRR
 
 | | X-RAY STRUCTURE OF THE BACTERIORHODOPSIN TRIMER/LIPID COMPLEX | | Descriptor: | 3,7,11,15-TETRAMETHYL-HEXADECAN-1-OL, 3-O-sulfo-beta-D-galactopyranose-(1-6)-alpha-D-mannopyranose-(1-2)-alpha-D-glucopyranose, GLYCEROL, ... | | Authors: | Essen, L.-O, Siegert, R, Oesterhelt, D. | | Deposit date: | 1998-07-28 | | Release date: | 1998-09-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Lipid patches in membrane protein oligomers: crystal structure of the bacteriorhodopsin-lipid complex
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
6F90
 
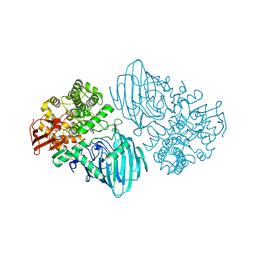 | | Structure of the family GH92 alpha-mannosidase BT3130 from Bacteroides thetaiotaomicron in complex with Mannoimidazole (ManI) | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, Alpha-1,2-mannosidase, putative, ... | | Authors: | Thompson, A.J, Spears, R.J, Zhu, Y, Suits, M.D.L, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2017-12-13 | | Release date: | 2018-05-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacteroides thetaiotaomicron generates diverse alpha-mannosidase activities through subtle evolution of a distal substrate-binding motif.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5FPU
 
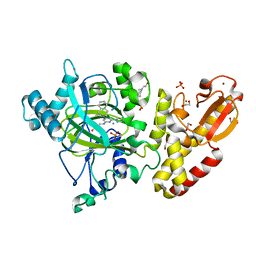 | | Crystal structure of human JARID1B in complex with GSKJ1 | | Descriptor: | 1,2-ETHANEDIOL, 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Srikannathasan, V, Nowak, R, Johansson, C, Gileadi, C, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Oppermann, U. | | Deposit date: | 2015-12-03 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Analysis of Human Kdm5B Guides Histone Demethylase Inhibitor Development.
Nat.Chem.Biol., 12, 2016
|
|
6AV2
 
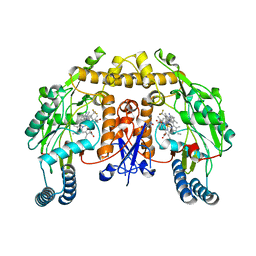 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 6-(3-(3-(dimethylamino)propyl)-5-(trifluoromethyl)phenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{3-[3-(dimethylamino)propyl]-5-(trifluoromethyl)phenyl}ethyl)-4-methylpyridin-2-amine, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Improvement of Cell Permeability of Human Neuronal Nitric Oxide Synthase Inhibitors Using Potent and Selective 2-Aminopyridine-Based Scaffolds with a Fluorobenzene Linker.
J. Med. Chem., 60, 2017
|
|
5TWZ
 
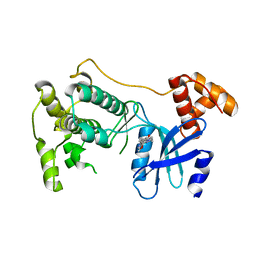 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 7-{[2-methoxy-4-(1H-pyrazol-4-yl)benzoyl]amino}-2,3,4,5-tetrahydro-1H-3-benzazepinium, Maternal embryonic leucine zipper kinase | | Authors: | Seo, H.-Y, Dhe-Paganon, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
8A0L
 
 | | Tubulin-CW1-complex | | Descriptor: | (3~{S},4~{R},8~{S},10~{S},12~{S},14~{S})-14-[(~{Z},4~{R})-4-(hydroxymethyl)hex-2-en-2-yl]-4,12-dimethoxy-9,9-dimethyl-3,8,10-tris(oxidanyl)-1-oxacyclotetradecan-2-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Prota, A.E, Diaz, J.F, Steinmetz, M.O, Oliva, M.A. | | Deposit date: | 2022-05-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9981 Å) | | Cite: | Chemical modulation of microtubule structure through the laulimalide/peloruside site.
Structure, 31, 2023
|
|
5TX5
 
 | | Rip1 Kinase ( flag 1-294, C34A, C127A, C233A, C240A) with GSK772 | | Descriptor: | 3-benzyl-N-[(3S)-5-methyl-4-oxo-2,3,4,5-tetrahydro-1,5-benzoxazepin-3-yl]-1H-1,2,4-triazole-5-carboxamide, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Campobasso, N, Ward, P, Thrope, J. | | Deposit date: | 2016-11-15 | | Release date: | 2017-07-05 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Discovery of a First-in-Class Receptor Interacting Protein 1 (RIP1) Kinase Specific Clinical Candidate (GSK2982772) for the Treatment of Inflammatory Diseases.
J. Med. Chem., 60, 2017
|
|
5FVS
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 4-methyl-6-(2-(5-(3-(methylamino)propyl)pyridin-3-yl) ethyl)pyridin-2-amine | | Descriptor: | 4-METHYL-6-(2-(5-(3-(METHYLAMINO)PROPYL)PYRIDIN-3-YL)ETHYL)PYRIDIN-2-AMINE, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2016-02-10 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Potent and Selective Human Neuronal Nitric Oxide Synthase Inhibition by Optimization of the 2-Aminopyridine-Based Scaffold with a Pyridine Linker.
J.Med.Chem., 59, 2016
|
|
5U1C
 
 | |
6AUT
 
 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 6-(3-(3-(Dimethylamino)propyl)-5-fluorophenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{3-[3-(dimethylamino)propyl]-5-fluorophenyl}ethyl)-4-methylpyridin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Improvement of Cell Permeability of Human Neuronal Nitric Oxide Synthase Inhibitors Using Potent and Selective 2-Aminopyridine-Based Scaffolds with a Fluorobenzene Linker.
J. Med. Chem., 60, 2017
|
|
1BWA
 
 | | HIV-1 PROTEASE (V82F/I84V) DOUBLE MUTANT COMPLEXED WITH XV638 OF DUPONT PHARMACEUTICALS | | Descriptor: | PROTEIN (HIV-1 PROTEASE), [4R-(4ALPHA,5ALPHA,6BETA,7BETA)]-3,3'-[[TETRAHYDRO-5,6-DIHYDROXY-2-OXO-4,7-BIS(PHENYLMETHYL)-1H-1,3-DIAZEPINE-1,3(2H)-D IYL] BIS(METHYLENE)]BIS[N-2-THIAZOLYLBENZAMIDE] | | Authors: | Ala, P, Chang, C.H. | | Deposit date: | 1998-09-22 | | Release date: | 1998-10-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Counteracting HIV-1 protease drug resistance: structural analysis of mutant proteases complexed with XV638 and SD146, cyclic urea amides with broad specificities.
Biochemistry, 37, 1998
|
|
5YAV
 
 | | Fragment-based Drug Discovery of inhibitors to block PDEdelta-RAS protein-protein interaction | | Descriptor: | 1-phenyl-5-propan-2-ylsulfanyl-1,2,3,4-tetrazole, Retinal rod rhodopsin-sensitive cGMP 3',5'-cyclic phosphodiesterase subunit delta | | Authors: | Bing, X, Yanlian, L, Danyan, C. | | Deposit date: | 2017-09-01 | | Release date: | 2018-09-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.989 Å) | | Cite: | Fragment-based Drug Discovery of inhibitors to block PDEdelta-RAS protein-protein interaction
To Be Published
|
|
5UEX
 
 | | BRD4_BD2_A-1497627 | | Descriptor: | 17-{[ethyl(dihydroxy)-lambda~4~-sulfanyl]amino}-11,13-difluoro-2-methyl-6,7,8,9-tetrahydrodibenzo[4,5:7,8][1,6]dioxacyclododecino[3,2-c]pyridin-3(2H)-one, Bromodomain-containing protein 4 | | Authors: | Park, C.H. | | Deposit date: | 2017-01-03 | | Release date: | 2017-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Fragment-Based, Structure-Enabled Discovery of Novel Pyridones and Pyridone Macrocycles as Potent Bromodomain and Extra-Terminal Domain (BET) Family Bromodomain Inhibitors.
J. Med. Chem., 60, 2017
|
|
7ZLJ
 
 | | Cryo-EM structure of C-mannosyltransferase CeDPY19, in ternary complex with Dol25-P-C-Man and acceptor peptide, bound to CMT2-Fab and anti-Fab nanobody | | Descriptor: | Anti-Fab nanobody, C-mannosyltransferase dpy-19, CMT2-Fab heavy chain, ... | | Authors: | Bloch, J.S, Mao, R, Mukherjee, S, Boilevin, J, Irobalieva, R, Darbre, T, Reymond, J.L, Kossiakoff, A.A, Goddard-Borger, E.D, Locher, K.P. | | Deposit date: | 2022-04-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Structure, sequon recognition and mechanism of tryptophan C-mannosyltransferase.
Nat.Chem.Biol., 19, 2023
|
|
