8ADK
 
 | |
8ADJ
 
 | | Poly(ADP-ribose) glycohydrolase (PARG) from Drosophila melanogaster in complex with PARG inhibitor PDD00017272 | | Descriptor: | 1-[(2,5-dimethylpyrazol-3-yl)methyl]-N-(1-methylcyclopropyl)-3-[(2-methyl-1,3-thiazol-5-yl)methyl]-2,4-bis(oxidanylidene)quinazoline-6-sulfonamide, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ariza, A, Fontana, P. | | Deposit date: | 2022-07-08 | | Release date: | 2023-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.508 Å) | | Cite: | Serine ADP-ribosylation in Drosophila provides insights into the evolution of reversible ADP-ribosylation signalling.
Nat Commun, 14, 2023
|
|
8ADI
 
 | | Cryo-EM structure of Darobactin 9 bound BAM complex | | Descriptor: | Darobactin 9, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Yuan, B, Marlovits, T.C. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Darobactins Exhibiting Superior Antibiotic Activity by Cryo-EM Structure Guided Biosynthetic Engineering.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8ADG
 
 | | Cryo-EM structure of Darobactin 22 bound BAM complex | | Descriptor: | Darobactin 22, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Yuan, B, Marlovits, T.C. | | Deposit date: | 2022-07-08 | | Release date: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Darobactins Exhibiting Superior Antibiotic Activity by Cryo-EM Structure Guided Biosynthetic Engineering.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8ADB
 
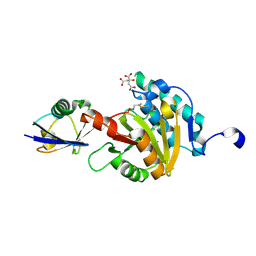 | | Viral tegument-like DUBs | | Descriptor: | CITRIC ACID, Polyubiquitin-C, Wc-VDT1, ... | | Authors: | Erven, I, Abraham, E.T, Hermanns, T, Baumann, U, Hofmann, K. | | Deposit date: | 2022-07-08 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A widely distributed family of eukaryotic and bacterial deubiquitinases related to herpesviral large tegument proteins.
Nat Commun, 13, 2022
|
|
8AD9
 
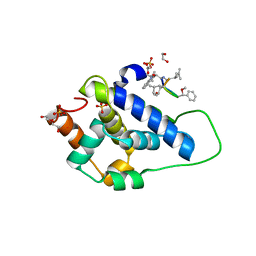 | | Crystal structure of ClpC2 C-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Cyclomarin A, ... | | Authors: | Taylor, G, Cui, H.J, Leodolter, J, Giese, C, Weber-Ban, E. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | ClpC2 protects mycobacteria against a natural antibiotic targeting ClpC1-dependent protein degradation.
Commun Biol, 6, 2023
|
|
8AD8
 
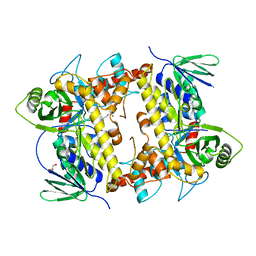 | | Flavin-dependent tryptophan 6-halogenase Thal in complex with a D-Trp-Ser dipeptide | | Descriptor: | D-tryptophyl-L-serine, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Gafe, S, Moritzer, A.C, Montua, N, Sewald, N, Niemann, H.H. | | Deposit date: | 2022-07-08 | | Release date: | 2022-10-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Enzymatic Late-Stage Halogenation of Peptides.
Chembiochem, 24, 2023
|
|
8AD7
 
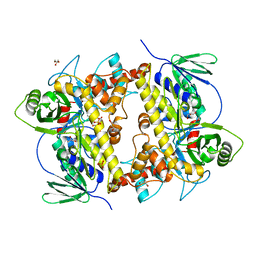 | |
8AD0
 
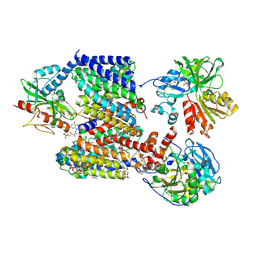 | |
8ACY
 
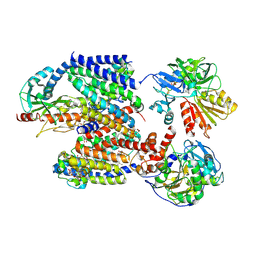 | | X-ray structure of Na+-NQR from Vibrio cholerae at 3.5 A resolution | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8ACW
 
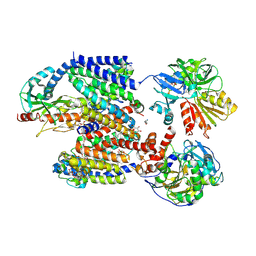 | | X-ray structure of Na+-NQR from Vibrio cholerae at 3.4 A resolution | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Fritz, G. | | Deposit date: | 2022-07-07 | | Release date: | 2023-07-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Conformational coupling of redox-driven Na + -translocation in Vibrio cholerae NADH:quinone oxidoreductase.
Nat.Struct.Mol.Biol., 30, 2023
|
|
8ACV
 
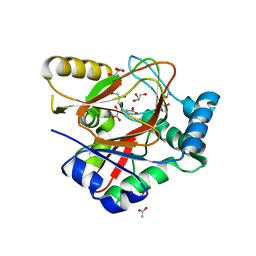 | | WelO5* bound to Zn(II), Cl, and 2-oxoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, CHLORIDE ION, ... | | Authors: | Buller, R, Hueppi, S, Voss, M, Hayashi, T. | | Deposit date: | 2022-07-07 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Enzyme engineering enables inversion of substrate stereopreference of the halogenase WelO5*
Chemcatchem, 2022
|
|
8ACS
 
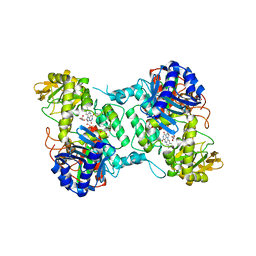 | | Crystal structure of FMO from Janthinobacterium svalbardensis | | Descriptor: | DI(HYDROXYETHYL)ETHER, FAD-dependent oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Polidori, N, Galuska, P, Gruber, K. | | Deposit date: | 2022-07-06 | | Release date: | 2022-09-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Cold-Active Flavin-Dependent Monooxygenase from Janthinobacterium svalbardensis Unlocks Applications of Baeyer-Villiger Monooxygenases at Low Temperature.
Acs Catalysis, 13, 2023
|
|
8ACQ
 
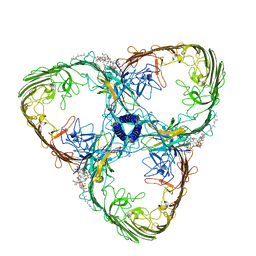 | | S-layer Deinoxanthin-Binding Complex (SDBC), subunit DR_2577 assembled with its SOD DR_0644 | | Descriptor: | (3~{S},5~{R},6~{R})-5-[(3~{S},7~{R},12~{S},16~{S},20~{S})-3,7,12,16,20,24-hexamethyl-24-oxidanyl-pentacosyl]-4,4,6-trimethyl-cyclohexane-1,3-diol, COPPER (II) ION, DR_0644, ... | | Authors: | Farci, D, Piano, D. | | Deposit date: | 2022-07-06 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | The SDBC is active in quenching oxidative conditions and bridges the cell envelope layers in Deinococcus radiodurans.
J.Biol.Chem., 299, 2023
|
|
8ACO
 
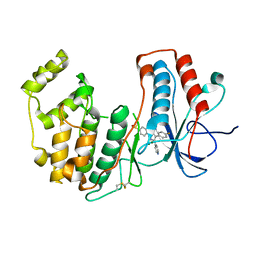 | | Crystal structure of WT p38alpha | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAGNESIUM ION, Mitogen-activated protein kinase 14 | | Authors: | Pous, J, Baginski, B, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of WT p38alpha
Res Sq
|
|
8ACM
 
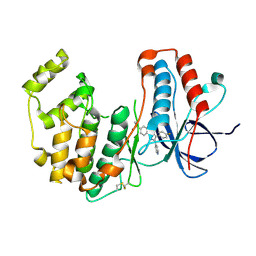 | | Crystal structure of WT p38alpha | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAGNESIUM ION, Mitogen-activated protein kinase 14 | | Authors: | Pous, J, Baginski, B, Gonzalez, L, Macias, M.J, Nebreda, A.R. | | Deposit date: | 2022-07-05 | | Release date: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of WT p38alpha
Res Sq
|
|
8ACL
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GC-14 | | Descriptor: | (2~{S})-1-(3,4-dichlorophenyl)-4-pyridin-3-ylcarbonyl-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
8ACK
 
 | |
8ACH
 
 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): large class from symmetry expansion | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.91 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
8ACD
 
 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the non-covalent inhibitor GA-17S | | Descriptor: | (2~{S})-4-[[2,4-bis(oxidanylidene)-1~{H}-pyrimidin-6-yl]carbonyl]-1-(3,4-dichlorophenyl)-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C.E, Sylvester, K, Claff, T, Weisse, R.H, Gao, S, Tollefson, A.E, Liu, X, Zhan, P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-09-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery and Crystallographic Studies of Trisubstituted Piperazine Derivatives as Non-Covalent SARS-CoV-2 Main Protease Inhibitors with High Target Specificity and Low Toxicity.
J.Med.Chem., 65, 2022
|
|
8AC7
 
 | |
8AC6
 
 | | Nudaurelia capensis omega virus maturation intermediate captured at pH5.6 (insect cell expressed VLPs): medium class from symmetry expansion | | Descriptor: | p70 | | Authors: | Castells-Graells, R, Hesketh, E.L, Johnson, J.E, Ranson, N.A, Lawson, D.M, Lomonossoff, G.P. | | Deposit date: | 2022-07-05 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Decoding virus maturation with cryo-EM structures of intermediates
To be published
|
|
8AC5
 
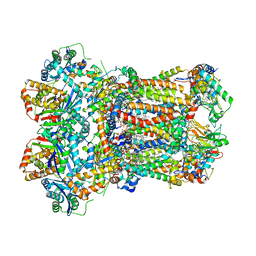 | | Complex III2 from Yarrowia lipolytica, with decylubiquinol, oxidised, b-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
8AC4
 
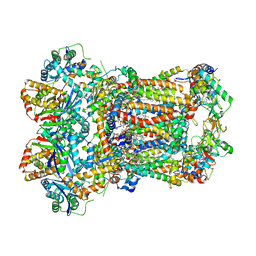 | | Complex III2 from Yarrowia lipolytica, apo, c-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
8AC3
 
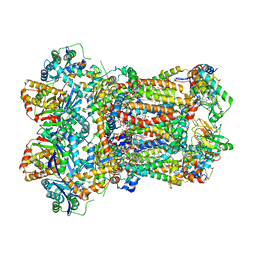 | | Complex III2 from Yarrowia lipolytica, apo, int-position | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHATE, CARDIOLIPIN, ... | | Authors: | Wieferig, J.P, Kuhlbrandt, W. | | Deposit date: | 2022-07-05 | | Release date: | 2023-01-11 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Analysis of the conformational heterogeneity of the Rieske iron-sulfur protein in complex III 2 by cryo-EM.
Iucrj, 10, 2023
|
|
