4WGL
 
 | | Crystal structure of a GroEL D83A/R197A double mutant | | Descriptor: | 60 kDa chaperonin | | Authors: | Yang, D, Fei, X, LaRonde, N.A, Beckett, D, Lund, P.A, Lorimer, G.H. | | Deposit date: | 2014-09-19 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.13 Å) | | Cite: | Crystal structure of a GroEL D83A/R197A double mutant
To Be Published
|
|
3RUS
 
 | | Crystal structure of Cpn-rls in complex with ADP from Methanococcus maripaludis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin, MAGNESIUM ION, ... | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N.R, Kumar, R, McAndrew, R.P, Knee, K.M, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2011-05-05 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.338 Å) | | Cite: | Mechanism of nucleotide sensing in group II chaperonins.
Embo J., 31, 2012
|
|
3AQ1
 
 | | Open state monomer of a group II chaperonin from methanococcoides burtonii | | Descriptor: | Thermosome subunit | | Authors: | Harrop, S.J, Pilak, O, Siddiqui, K.S, De Francisci, D, Burg, D, Williams, T.J, Cavicchioli, R, Curmi, P.M. | | Deposit date: | 2010-10-24 | | Release date: | 2011-09-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Chaperonins from an Antarctic archaeon are predominantly monomeric: crystal structure of an open state monomer.
ENVIRON.MICROBIOL., 13, 2011
|
|
4WSC
 
 | | Crystal structure of a GroELK105A mutant | | Descriptor: | 60 kDa chaperonin | | Authors: | Lorimer, G.H, Ye, X, Fei, X, Yang, D, Corsepius, N, LaRonde, N.A. | | Deposit date: | 2014-10-26 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Crystal structure of a GroELK105A mutant
To Be Published
|
|
1LA1
 
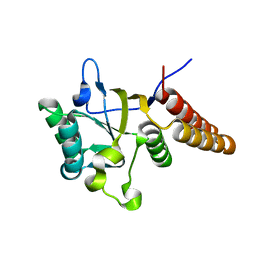 | | Gro-EL Fragment (Apical Domain) Comprising Residues 188-379 | | Descriptor: | GroEL | | Authors: | Ashcroft, A.E, Brinker, A, Coyle, J.E, Weber, F, Kaiser, M, Moroder, L, Parsons, M.R, Jager, J, Hartl, U.F, Hayer-Hartl, M, Radford, S.E. | | Deposit date: | 2002-03-27 | | Release date: | 2002-04-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural plasticity and noncovalent substrate binding in the GroEL apical domain. A study using electrospay ionization mass spectrometry and fluorescence binding studies.
J.Biol.Chem., 277, 2002
|
|
4XCG
 
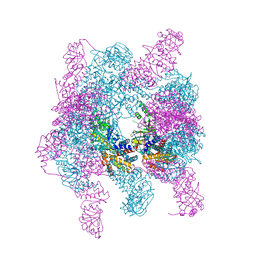 | | Crystal structure of a hexadecameric TF55 complex from S. solfataricus, crystal form I | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Thermosome subunit alpha, Thermosome subunit beta | | Authors: | Stewart, A.G, Smits, C, Chaston, J.J, Stock, D. | | Deposit date: | 2014-12-18 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.737 Å) | | Cite: | Structural and Functional Insights into the Evolution and Stress Adaptation of Type II Chaperonins.
Structure, 24, 2016
|
|
1IOK
 
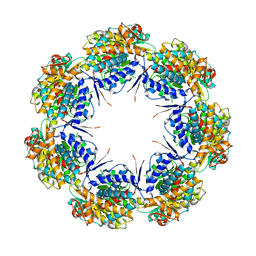 | | CRYSTAL STRUCTURE OF CHAPERONIN-60 FROM PARACOCCUS DENITRIFICANS | | Descriptor: | CHAPERONIN 60 | | Authors: | Fukami, T.A, Yohda, M, Taguchi, H, Yoshida, M, Miki, K. | | Deposit date: | 2001-03-16 | | Release date: | 2001-10-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of chaperonin-60 from Paracoccus denitrificans.
J.Mol.Biol., 312, 2001
|
|
3CAU
 
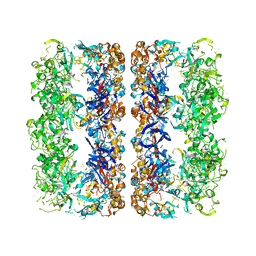 | | D7 symmetrized structure of unliganded GroEL at 4.2 Angstrom resolution by cryoEM | | Descriptor: | 60 kDa chaperonin | | Authors: | Ludtke, S.J, Baker, M.L, Chen, D.H, Song, J.L, Chuang, D, Chiu, W. | | Deposit date: | 2008-02-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | De Novo Backbone Trace of GroEL from Single Particle Electron Cryomicroscopy.
Structure, 16, 2008
|
|
3C9V
 
 | | C7 Symmetrized Structure of Unliganded GroEL at 4.7 Angstrom Resolution from CryoEM | | Descriptor: | 60 kDa chaperonin | | Authors: | Ludtke, S.J, Baker, M.L, Chen, D.H, Song, J.L, Chuang, D, Chiu, W. | | Deposit date: | 2008-02-18 | | Release date: | 2008-09-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | De Novo Backbone Trace of GroEL from Single Particle Electron Cryomicroscopy.
Structure, 16, 2008
|
|
2EU1
 
 | | Crystal structure of the chaperonin GroEL-E461K | | Descriptor: | GROEL | | Authors: | Cabo-Bilbao, A, Spinelli, S, Sot, B, Agirre, J, Mechaly, A.E, Muga, A, Guerin, D.M.A. | | Deposit date: | 2005-10-28 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Crystal structure of the temperature-sensitive and allosteric-defective chaperonin GroEL(E461K).
J.Struct.Biol., 155, 2006
|
|
7NVL
 
 | | Human TRiC complex in closed state with nanobody bound (Consensus Map) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Kelly, J.J, Chi, G, Bulawa, C, Paavilainen, V.O, Bountra, C, Huiskonen, J.T, Yue, W, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-02 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Snapshots of actin and tubulin folding inside the TRiC chaperonin.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7NVO
 
 | | Human TRiC complex in open state with nanobody bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Kelly, J.J, Chi, G, Bulawa, C, Paavilainen, V.O, Bountra, C, Huiskonen, J.T, Yue, W. | | Deposit date: | 2021-03-15 | | Release date: | 2022-03-02 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Snapshots of actin and tubulin folding inside the TRiC chaperonin.
Nat.Struct.Mol.Biol., 29, 2022
|
|
1ASX
 
 | | APICAL DOMAIN OF THE CHAPERONIN FROM THERMOPLASMA ACIDOPHILUM | | Descriptor: | PHOSPHATE ION, THERMOSOME | | Authors: | Klumpp, M, Baumeister, W, Essen, L.-O. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the substrate binding domain of the thermosome, an archaeal group II chaperonin.
Cell(Cambridge,Mass.), 91, 1997
|
|
1ASS
 
 | | APICAL DOMAIN OF THE CHAPERONIN FROM THERMOPLASMA ACIDOPHILUM | | Descriptor: | PHOSPHATE ION, SODIUM ION, THERMOSOME | | Authors: | Klumpp, M, Baumeister, W, Essen, L.-O. | | Deposit date: | 1997-08-11 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the substrate binding domain of the thermosome, an archaeal group II chaperonin.
Cell(Cambridge,Mass.), 91, 1997
|
|
1A6E
 
 | | THERMOSOME-MG-ADP-ALF3 COMPLEX | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Ditzel, L, Loewe, J, Stock, D, Stetter, K.-O, Huber, H, Huber, R, Steinbacher, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT.
Cell(Cambridge,Mass.), 93, 1998
|
|
1A6D
 
 | | THERMOSOME FROM T. ACIDOPHILUM | | Descriptor: | THERMOSOME (ALPHA SUBUNIT), THERMOSOME (BETA SUBUNIT) | | Authors: | Ditzel, L, Loewe, J, Stock, D, Stetter, K.-O, Huber, H, Huber, R, Steinbacher, S. | | Deposit date: | 1998-02-24 | | Release date: | 1999-03-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the thermosome, the archaeal chaperonin and homolog of CCT.
Cell(Cambridge,Mass.), 93, 1998
|
|
1SS8
 
 | | GroEL | | Descriptor: | groEL protein | | Authors: | Chaudhry, C, Horwich, A.L, Brunger, A.T, Adams, P.D. | | Deposit date: | 2004-03-23 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Exploring the structural dynamics of the E.coli chaperonin GroEL using translation-libration-screw crystallographic refinement of intermediate states.
J.Mol.Biol., 342, 2004
|
|
1SRV
 
 | | THERMUS THERMOPHILUS GROEL (HSP60 CLASS) FRAGMENT (APICAL DOMAIN) COMPRISING RESIDUES 192-336 | | Descriptor: | PROTEIN (GROEL (HSP60 CLASS)) | | Authors: | Walsh, M.A, Dementieva, I, Evans, G, Sanishvili, R, Joachimiak, A. | | Deposit date: | 1999-03-02 | | Release date: | 1999-03-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Taking MAD to the extreme: ultrafast protein structure determination.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1SJP
 
 | |
1SX3
 
 | | GroEL14-(ATPgammaS)14 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Chaudhry, C, Horwich, A.L, Brunger, A.T, Adams, P.D. | | Deposit date: | 2004-03-30 | | Release date: | 2005-03-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploring the structural dynamics of the E.coli chaperonin GroEL using translation-libration-screw crystallographic refinement of intermediate states.
J.Mol.Biol., 342, 2004
|
|
1E0R
 
 | | Beta-apical domain of thermosome | | Descriptor: | THERMOSOME | | Authors: | Bosch, G, Baumeister, W, Essen, L.-O. | | Deposit date: | 2000-04-06 | | Release date: | 2000-08-19 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Beta-Apical Domain from Thermosome Reveals Structural Plasticity in Protrusion Region
J.Mol.Biol., 301, 2000
|
|
8G7M
 
 | | ATP-bound mtHsp60 V72I focus | | Descriptor: | 60 kDa heat shock protein, mitochondrial, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7L
 
 | | ATP-bound mtHsp60 V72I | | Descriptor: | 60 kDa heat shock protein, mitochondrial, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7J
 
 | | mtHsp60 V72I apo | | Descriptor: | 60 kDa heat shock protein, mitochondrial | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
8G7K
 
 | | mtHsp60 V72I apo focus | | Descriptor: | 60 kDa heat shock protein, mitochondrial | | Authors: | Braxton, J.R, Shao, H, Tse, E, Gestwicki, J.E, Southworth, D.R. | | Deposit date: | 2023-02-16 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Asymmetric apical domain states of mitochondrial Hsp60 coordinate substrate engagement and chaperonin assembly.
Biorxiv, 2023
|
|
