3RBS
 
 | |
1NDA
 
 | | THE STRUCTURE OF TRYPANOSOMA CRUZI TRYPANOTHIONE REDUCTASE IN THE OXIDIZED AND NADPH REDUCED STATE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, TRYPANOTHIONE OXIDOREDUCTASE | | Authors: | Lantwin, C.B, Kabsch, W, Pai, E.F, Schlichting, I, Krauth-Siegel, R.L. | | Deposit date: | 1993-07-02 | | Release date: | 1994-09-30 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of Trypanosoma cruzi trypanothione reductase in the oxidized and NADPH reduced state.
Proteins, 18, 1994
|
|
2CAN
 
 | |
1IX6
 
 | | Aspartate Aminotransferase Active Site Mutant V39F | | Descriptor: | Aspartate Aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Hayashi, H, Mizuguchi, H, Miyahara, I, Nakajima, Y, Hirotsu, K, Kagamiyama, H. | | Deposit date: | 2002-06-14 | | Release date: | 2002-07-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational change in aspartate aminotransferase on substrate binding induces strain in the catalytic group and enhances catalysis
J.BIOL.CHEM., 278, 2003
|
|
3VOM
 
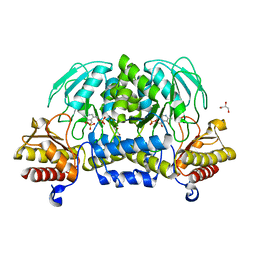 | | Structure of a putative phosphoserine aminotransferase from mycobacterium tuberculosis | | Descriptor: | GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, Putative phosphoserine aminotransferase, ... | | Authors: | Coulibaly, F, Lassalle, E, Baker, H.M, Baker, E.N. | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of phosphoserine aminotransferase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1E4U
 
 | | N-terminal RING finger domain of human NOT-4 | | Descriptor: | TRANSCRIPTIONAL REPRESSOR NOT4, ZINC ION | | Authors: | Hanzawa, H, De Ruwe, M.J, Albert, T.K, Van Der Vliet, P.C, Timmers, H.T, Boelens, R. | | Deposit date: | 2000-07-12 | | Release date: | 2001-03-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of the C4C4 Ring Finger of Human not4 Reveals Features Distinct from Those of C3Hc4 Ring Fingers
J.Biol.Chem., 276, 2001
|
|
3EI8
 
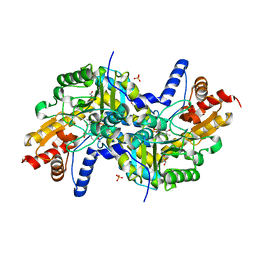 | | Crystal structure of K270N variant of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with LL-DAP: External aldimine form | | Descriptor: | (2S,6S)-2-amino-6-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}heptanedioic acid, GLYCEROL, LL-diaminopimelate aminotransferase, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3EI5
 
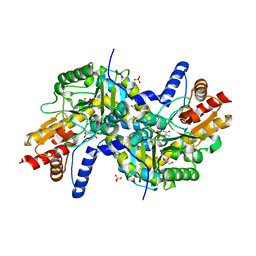 | | Crystal structure of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with PLP-Glu: an external aldimine mimic | | Descriptor: | GLYCEROL, LL-diaminopimelate aminotransferase, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-glutamic acid, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3EIA
 
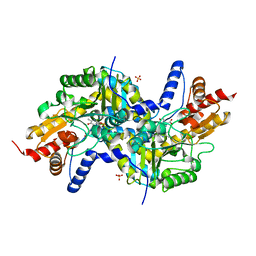 | | Crystal structure of K270Q variant of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with L-Glu: External aldimine form | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-glutamic acid, LL-diaminopimelate aminotransferase, SULFATE ION | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3EI7
 
 | | Crystal structure of apo-LL-diaminopimelate aminotransferase from Arabidopsis thaliana (no PLP) | | Descriptor: | LL-diaminopimelate aminotransferase, SULFATE ION | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3ZZK
 
 | | Structure of an engineered aspartate aminotransferase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, GLYCEROL, ... | | Authors: | Fernandez, F.J, deVries, D, Pena-Soler, E, Coll, M, Christen, P, Gehring, H, Vega, M.C. | | Deposit date: | 2011-09-01 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure and Mechanism of a Cysteine Sulfinate Desulfinase Engineered on the Aspartate Aminotransferase Scaffold.
Biocim.Biophys.Acta, 1824, 2011
|
|
3ZZJ
 
 | | Structure of an engineered aspartate aminotransferase | | Descriptor: | ASPARTATE AMINOTRANSFERASE, BETA-MERCAPTOETHANOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fernandez, F.J, deVries, D, Pena-Soler, E, Coll, M, Christen, P, Gehring, H, Vega, M.C. | | Deposit date: | 2011-09-01 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure and Mechanism of a Cysteine Sulfinate Desulfinase Engineered on the Aspartate Aminotransferase Scaffold.
Biocim.Biophys.Acta, 1824, 2011
|
|
3UYY
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
3EI9
 
 | | Crystal structure of K270N variant of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with L-Glu: External aldimine form | | Descriptor: | (E)-N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-glutamic acid, GLYCEROL, LL-diaminopimelate aminotransferase, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
2FYF
 
 | | Structure of a putative phosphoserine aminotransferase from Mycobacterium Tuberculosis | | Descriptor: | GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, SULFATE ION, ... | | Authors: | Coulibaly, F, Lassalle, E, Baker, E.N, Mycobacterium Tuberculosis Structural Proteomics Project (XMTB) | | Deposit date: | 2006-02-07 | | Release date: | 2007-01-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of phosphoserine aminotransferase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
3UZB
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, Branched-chain-amino-acid aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
3EIB
 
 | | Crystal structure of K270N variant of LL-diaminopimelate aminotransferase from Arabidopsis thaliana | | Descriptor: | GLYCEROL, LL-diaminopimelate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
3EI6
 
 | | Crystal structure of LL-diaminopimelate aminotransferase from Arabidopsis thaliana complexed with PLP-DAP: an external aldimine mimic | | Descriptor: | (2S,6S)-2-amino-6-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)amino]heptanedioic acid, GLYCEROL, LL-diaminopimelate aminotransferase, ... | | Authors: | Watanabe, N, Clay, M.D, van Belkum, M.J, Cherney, M.M, Vederas, J.C, James, M.N.G. | | Deposit date: | 2008-09-15 | | Release date: | 2008-10-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of substrate recognition and PLP-induced conformational changes in LL-diaminopimelate aminotransferase from Arabidopsis thaliana.
J.Mol.Biol., 384, 2008
|
|
4A00
 
 | | Structure of an engineered aspartate aminotransferase | | Descriptor: | ALANYL-PYRIDOXAL-5'-PHOSPHATE, ASPARTATE AMINOTRANSFERASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Fernandez, F.J, deVries, D, Pena-Soler, E, Coll, M, Christen, P, Gehring, H, Vega, M.C. | | Deposit date: | 2011-09-06 | | Release date: | 2011-12-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structure and Mechanism of a Cysteine Sulfinate Desulfinase Engineered on the Aspartate Aminotransferase Scaffold.
Biocim.Biophys.Acta, 1824, 2011
|
|
3UZO
 
 | | Crystal Structures of Branched-Chain Aminotransferase from Deinococcus radiodurans Complexes with alpha-Ketoisocaproate and L-Glutamate Suggest Its Radio-Resistance for Catalysis | | Descriptor: | Branched-chain-amino-acid aminotransferase, GLUTAMIC ACID, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Chen, C.D, Huang, Y.C, Chuankhayan, P, Hsieh, Y.C, Huang, T.F, Lin, C.H, Guan, H.H, Liu, M.Y, Chang, W.C, Chen, C.J. | | Deposit date: | 2011-12-07 | | Release date: | 2012-12-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Complexes of the Branched-Chain Aminotransferase from Deinococcus radiodurans with alpha-Ketoisocaproate and L-Glutamate Suggest the Radiation Resistance of This Enzyme for Catalysis
J.Bacteriol., 194, 2012
|
|
1BWD
 
 | | INOSAMINE-PHOSPHATE AMIDINOTRANSFERASE STRB1 FROM STREPTOMYCES GRISEUS | | Descriptor: | PROTEIN (INOSAMINE-PHOSPHATE AMIDINOTRANSFERASE) | | Authors: | Fritsche, E, Bergner, A, Humm, A, Piepersberg, W, Huber, R. | | Deposit date: | 1998-09-23 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of L-arginine:inosamine-phosphate amidinotransferase StrB1 from Streptomyces griseus: an enzyme involved in streptomycin biosynthesis.
Biochemistry, 37, 1998
|
|
1KAJ
 
 | |
1SZK
 
 | | The structure of gamma-aminobutyrate aminotransferase mutant: E211S | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-04-05 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
3LG0
 
 | |
1SZS
 
 | | The structure of gamma-aminobutyrate aminotransferase mutant: I50Q | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Zhou, X, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-04-06 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
