1LNH
 
 | |
2K7H
 
 | | NMR solution structure of soybean allergen Gly m 4 | | Descriptor: | Stress-induced protein SAM22 | | Authors: | Berkner, H, Neudecker, P, Mittag, D, Ballmer-Weber, B.K, Schweimer, K, Vieths, S, Roesch, P. | | Deposit date: | 2008-08-11 | | Release date: | 2009-05-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Cross-reactivity of pollen and food allergens: soybean Gly m 4 is a member of the Bet v 1 superfamily and closely resembles yellow lupine proteins
Biosci.Rep., 29, 2009
|
|
1ROV
 
 | | Lipoxygenase-3 Treated with Cumene Hydroperoxide | | Descriptor: | FE (III) ION, Seed lipoxygenase-3 | | Authors: | Vahedi-Faridi, A, Brault, P.A, Shah, P, Kim, Y.W, Dunham, W.R, Funk, M.O. | | Deposit date: | 2003-12-02 | | Release date: | 2004-03-16 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Interaction between non-heme iron of lipoxygenases and cumene hydroperoxide: basis for enzyme activation, inactivation, and inhibition
J.Am.Chem.Soc., 126, 2004
|
|
2KN2
 
 | |
2KSZ
 
 | |
1S6J
 
 | |
1NO3
 
 | | REFINED STRUCTURE OF SOYBEAN LIPOXYGENASE-3 WITH 4-NITROCATECHOL AT 2.15 ANGSTROM RESOLUTION | | Descriptor: | 4-NITROCATECHOL, FE (III) ION, Lipoxygenase-3 | | Authors: | Skrzypczak-Jankun, E, Borbulevych, O.Y, Jankun, J. | | Deposit date: | 2003-01-15 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Soybean lipoxygenase-3 in complex with 4-nitrocatechol.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1S6I
 
 | |
1FMH
 
 | |
2L1W
 
 | |
2VCF
 
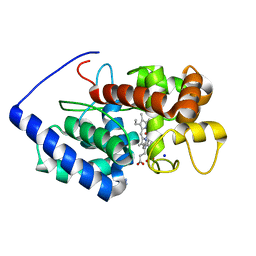 | | Structure of isoniazid (INH) bound to cytosolic soybean ascorbate peroxidase | | Descriptor: | 4-(DIAZENYLCARBONYL)PYRIDINE, ASCORBATE PEROXIDASE FROM SOYBEAN CYTOSOL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Metcalfe, C.L, Macdonald, I.K, Brown, K.A, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2007-09-21 | | Release date: | 2007-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Tuberculosis Prodrug Isoniazid Bound to Activating Peroxidases.
J.Biol.Chem., 283, 2008
|
|
1UIK
 
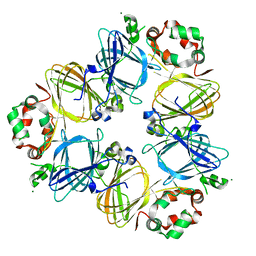 | | Crystal structure of soybean beta-conglycinin alpha prime homotrimer | | Descriptor: | MAGNESIUM ION, alpha prime subunit of beta-conglycinin | | Authors: | Maruyama, Y, Maruyama, N, Mikami, B, Utsumi, S. | | Deposit date: | 2003-07-16 | | Release date: | 2004-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the core region of the soybean beta-conglycinin alpha' subunit.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1UIJ
 
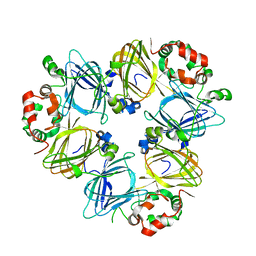 | | Crystal Structure Of Soybean beta-Conglycinin Beta Homotrimer (I122M/K124W) | | Descriptor: | beta subunit of beta conglycinin | | Authors: | Maruyama, N, Maruyama, Y, Tsuruki, T, Okuda, E, Yoshikawa, M, Utsumi, S. | | Deposit date: | 2003-07-16 | | Release date: | 2004-07-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Creation of soybean beta-conglycinin beta with strong phagocytosis-stimulating activity
BIOCHIM.BIOPHYS.ACTA, 1648, 2003
|
|
1UKP
 
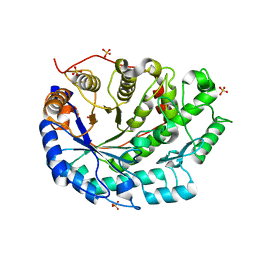 | | Crystal structure of soybean beta-amylase mutant substituted at surface region | | Descriptor: | Beta-amylase, SULFATE ION | | Authors: | Kang, Y.N, Adachi, M, Mikami, B, Utsumi, S. | | Deposit date: | 2003-08-31 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Change in the crystal packing of soybean beta-amylase mutants substituted at a few surface amino acid residues
Protein Eng., 16, 2003
|
|
1UKO
 
 | | Crystal structure of soybean beta-amylase mutant substituted at surface region | | Descriptor: | Beta-amylase, SULFATE ION | | Authors: | Kang, Y.N, Adachi, M, Mikami, B, Utsumi, S. | | Deposit date: | 2003-08-30 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Change in the crystal packing of soybean beta-amylase mutants substituted at a few surface amino acid residues
Protein Eng., 16, 2003
|
|
1V3H
 
 | | The roles of Glu186 and Glu380 in the catalytic reaction of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2003-11-02 | | Release date: | 2004-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Roles of Glu186 and Glu380 in the Catalytic Reaction of Soybean beta-Amylase.
J.Mol.Biol., 339, 2004
|
|
1WDS
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose, ... | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
1WDP
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
1WDQ
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
1WDR
 
 | | The role of an inner loop in the catalytic mechanism of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose, ... | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2004-05-17 | | Release date: | 2005-04-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural analysis of threonine 342 mutants of soybean beta-amylase: role of a conformational change of the inner loop in the catalytic mechanism.
Biochemistry, 44, 2005
|
|
1UWZ
 
 | | Bacillus subtilis cytidine deaminase with an Arg56 - Ala substitution | | Descriptor: | CYTIDINE DEAMINASE, TETRAHYDRODEOXYURIDINE, ZINC ION | | Authors: | Johansson, E, Neuhard, J, Willemoes, M, Larsen, S. | | Deposit date: | 2004-02-18 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural, Kinetic, and Mutational Studies of the Zinc Ion Environment in Tetrameric Cytidine Deaminase
Biochemistry, 43, 2004
|
|
1UX0
 
 | | Bacillus subtilis cytidine deaminase with an Arg56 - Gln substitution | | Descriptor: | CYTIDINE DEAMINASE, TETRAHYDRODEOXYURIDINE, ZINC ION | | Authors: | Johansson, E, Neuhard, J, Willemoes, M, Larsen, S. | | Deposit date: | 2004-02-18 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural, Kinetic, and Mutational Studies of the Zinc Ion Environment in Tetrameric Cytidine Deaminase
Biochemistry, 43, 2004
|
|
1V3I
 
 | | The roles of Glu186 and Glu380 in the catalytic reaction of soybean beta-amylase | | Descriptor: | Beta-amylase, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Kang, Y.N, Adachi, M, Utsumi, S, Mikami, B. | | Deposit date: | 2003-11-02 | | Release date: | 2004-06-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Roles of Glu186 and Glu380 in the Catalytic Reaction of Soybean beta-Amylase.
J.Mol.Biol., 339, 2004
|
|
7TN6
 
 | |
7TN5
 
 | |
