7PF6
 
 | |
7PF2
 
 | |
7PFD
 
 | |
7PF0
 
 | |
4FCX
 
 | | S.pombe Mre11 apoenzym | | Descriptor: | DNA repair protein rad32, MANGANESE (II) ION | | Authors: | Schiller, C.B, Lammens, K, Hopfner, K.P. | | Deposit date: | 2012-05-25 | | Release date: | 2012-06-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Mre11-Nbs1 complex yields insights into ataxia-telangiectasia-like disease mutations and DNA damage signaling.
Nat.Struct.Mol.Biol., 19, 2012
|
|
2VXC
 
 | |
6RNY
 
 | | PFV intasome - nucleosome strand transfer complex | | Descriptor: | DNA (108-MER), DNA (128-MER), DNA (33-MER), ... | | Authors: | Pye, V.E, Renault, L, Maskell, D.P, Cherepanov, P, Costa, A. | | Deposit date: | 2019-05-09 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Retroviral integration into nucleosomes through DNA looping and sliding along the histone octamer.
Nat Commun, 10, 2019
|
|
1CW9
 
 | | DNA DECAMER WITH AN ENGINEERED CROSSLINK IN THE MINOR GROOVE | | Descriptor: | 5'-D(*CP*CP*AP*GP*(G47)P*CP*CP*TP*GP*G)-3', CALCIUM ION | | Authors: | van Aalten, D.M.F, Erlanson, D.A, Verdine, G.L, Joshua-Tor, L. | | Deposit date: | 1999-08-26 | | Release date: | 1999-10-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | A structural snapshot of base-pair opening in DNA.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
2VXB
 
 | | Structure of the Crb2-BRCT2 domain | | Descriptor: | DNA REPAIR PROTEIN RHP9, PRASEODYMIUM ION | | Authors: | Kilkenny, M.L, Roe, S.M, Pearl, L.H. | | Deposit date: | 2008-07-03 | | Release date: | 2008-08-12 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Analysis of the Crb2-Brct2 Domain Reveals Distinct Roles in Checkpoint Signaling and DNA Damage Repair.
Genes Dev., 22, 2008
|
|
5UJY
 
 | |
5WJR
 
 | |
5U6Z
 
 | |
3PT9
 
 | | Crystal structure of mouse DNMT1(731-1602) in the free state | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Song, J, Patel, D.J. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation.
Science, 331, 2011
|
|
7PFX
 
 | |
7PFC
 
 | |
7PEY
 
 | |
7PEX
 
 | |
7PEZ
 
 | |
7PET
 
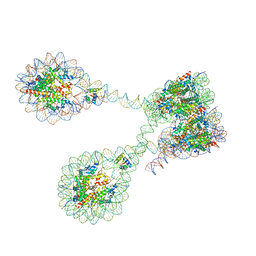 | | The 4x177 nucleosome array containing H1 | | Descriptor: | DNA (702-MER), Histone H1.4, Histone H2A type 1-B/E, ... | | Authors: | Dombrowski, M, Cramer, P. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Histone H1 binding to nucleosome arrays depends on linker DNA length and trajectory.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PF3
 
 | |
7PFU
 
 | |
7PFA
 
 | |
7PEU
 
 | |
7PFE
 
 | |
7PFT
 
 | |
