3DU3
 
 | | E(L212)A, D(L213)A, A(M249)Y triple mutant structure of photosynthetic reaction center | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CARDIOLIPIN, ... | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2008-07-16 | | Release date: | 2009-06-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | E(L212)A, D(L213)A, A(M249)Y triple mutant structure of photosynthetic reaction center
To be Published
|
|
3DSY
 
 | |
3DTS
 
 | |
3DU2
 
 | |
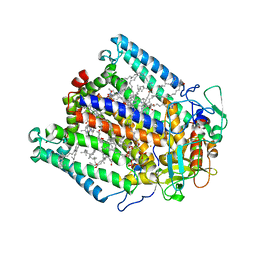
3DTR
 
 | |
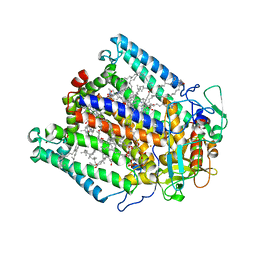
3DTA
 
 | |
5B5M
 
 | | Crystal structure of the Sr-substituted LH1-RC complex from Tch. tepidum | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Wang-Otomo, Z.-Y, Yu, L.-J. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis for the Unusual Qy Red-Shift and Enhanced Thermostability of the LH1 Complex from Thermochromatium tepidum.
Biochemistry, 55, 2016
|
|
7VY2
 
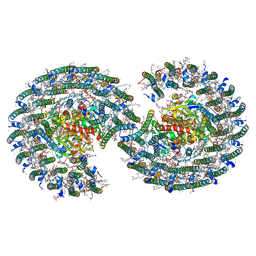 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF RHODOBACTER SPHAEROIDES DIMER | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Antenna pigment protein alpha chain, Antenna pigment protein beta chain, ... | | Authors: | Tani, K, Kanno, R, Kawamura, S, Kikuchi, R, Nagashima, K.V.P, Hall, M, Takahashi, A, Yu, L.-J, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2021-11-13 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Asymmetric structure of the native Rhodobacter sphaeroides dimeric LH1-RC complex.
Nat Commun, 13, 2022
|
|
5B5N
 
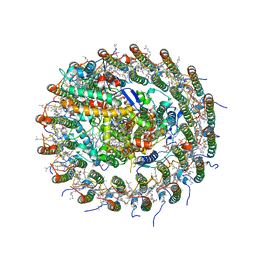 | | Crystal structure of the Ba-substituted LH1-RC complex from Tch. tepidum | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, ... | | Authors: | Wang-Otomo, Z.-Y, Yu, L.-J. | | Deposit date: | 2016-05-12 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis for the Unusual Qy Red-Shift and Enhanced Thermostability of the LH1 Complex from Thermochromatium tepidum.
Biochemistry, 55, 2016
|
|
1UMX
 
 | | PHOTOSYNTHETIC REACTION CENTER MUTANT WITH ARG M267 REPLACED WITH LEU (CHAIN M, R267L) | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN B, FE (III) ION, ... | | Authors: | Fyfe, P.K, Isaacs, N.W, Cogdell, R.J, Jones, M.R. | | Deposit date: | 2003-09-02 | | Release date: | 2004-06-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Disruption of a specific molecular interaction with a bound lipid affects the thermal stability of the purple bacterial reaction centre.
Biochim.Biophys.Acta, 1608, 2004
|
|
7F0L
 
 | | STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF RHODOBACTER SPHAEROIDES MONOMER | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, Antenna pigment protein beta chain, BACTERIOCHLOROPHYLL A, ... | | Authors: | Tani, K, Nagashima, V.P, Kanno, R, Kawamura, S, Kikuchi, R, Ji, X.-C, Hall, M, Yu, L.-J, Kimura, Y, Madigan, M.T, Mizoguchi, A, Humbel, B.M, Wang-Otomo, Z.-Y. | | Deposit date: | 2021-06-05 | | Release date: | 2021-11-10 | | Last modified: | 2021-11-17 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | A previously unrecognized membrane protein in the Rhodobacter sphaeroides LH1-RC photocomplex.
Nat Commun, 12, 2021
|
|
3ZUW
 
 | | Photosynthetic Reaction Centre Mutant with TYR L128 replaced with HIS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Gibasiewicz, K, Pajzderska, M, Potter, J.A, Fyfe, P.K, Dobek, A, Brettel, K, Jones, M.R. | | Deposit date: | 2011-07-20 | | Release date: | 2011-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Mechanism of Recombination of the P(+)H(A)(-) Radical Pair in Mutant Rhodobacter Sphaeroides Reaction Centers with Modified Free Energy Gaps between P(+)B(A)(-) and P(+)H(A)(-).
J Phys Chem B, 115, 2011
|
|
6RDJ
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1A, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDU
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1E, monomer-masked refinement | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6REC
 
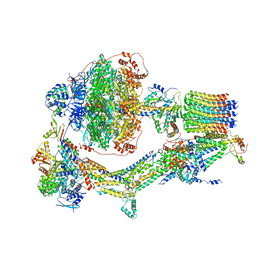 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 3A, monomer-masked refinement | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDA
 
 | | CryoEM structure of Polytomella F-ATP synthase, Primary rotary state 1, monomer-masked refinement | | Descriptor: | ASA-10: Polytomella F-ATP synthase associated subunit 10, ASA-2: Polytomella F-ATP synthase associated subunit 2, ASA-9: Polytomella F-ATP synthase associated subunit 9, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDD
 
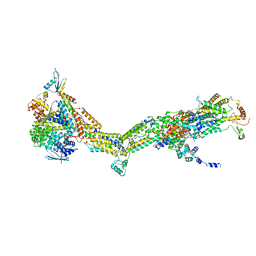 | | Cryo-EM structure of Polytomella F-ATP synthase, Primary rotary state 2, monomer-masked refinement | | Descriptor: | ASA-10: Polytomella F-ATP synthase associated subunit 10, ATP synthase associated protein ASA1, ATP synthase subunit alpha, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDN
 
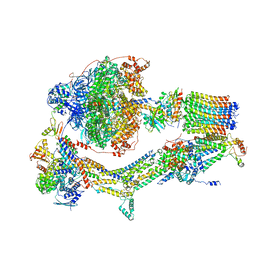 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1C, monomer-masked refinement | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RE3
 
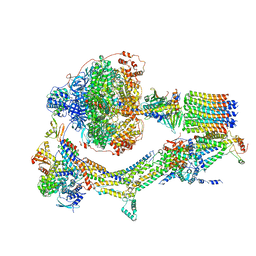 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2B, monomer-masked refinement | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6REU
 
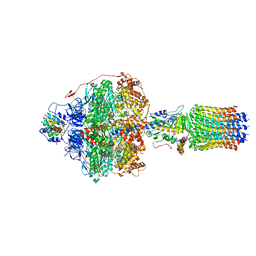 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 3C, focussed refinement of F1 head and rotor | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase gamma chain, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDK
 
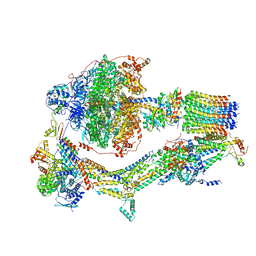 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1B, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RE0
 
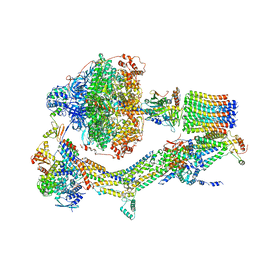 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 2A, monomer-masked refinement | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6REP
 
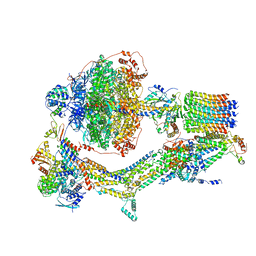 | | Cryo-EM structure of Polytomella F-ATP synthase, Primary rotary state 3, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RD9
 
 | | CryoEM structure of Polytomella F-ATP synthase, Primary rotary state 1, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
6RDT
 
 | | Cryo-EM structure of Polytomella F-ATP synthase, Rotary substate 1E, composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ASA-10: Polytomella F-ATP synthase associated subunit 10, ... | | Authors: | Murphy, B.J, Klusch, N, Yildiz, O, Kuhlbrandt, W. | | Deposit date: | 2019-04-12 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Rotary substates of mitochondrial ATP synthase reveal the basis of flexible F 1 -F o coupling.
Science, 364, 2019
|
|
