7ZMG
 
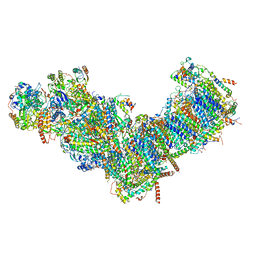 | | CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 1) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, Acyl carrier protein, ... | | Authors: | Laube, E, Kuehlbrandt, W. | | Deposit date: | 2022-04-19 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Conformational changes in mitochondrial complex I of the thermophilic eukaryote Chaetomium thermophilum.
Sci Adv, 8, 2022
|
|
7QSX
 
 | | Non-obligately L8S8-complex forming RubisCO derived from ancestral sequence reconstruction and rational engineering in L8 complex | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
7QT1
 
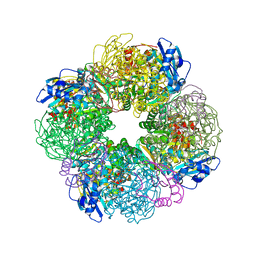 | | Non-obligately L8S8-complex forming RubisCO derived from ancestral sequence reconstruction and rational engineering in L8S8 complex with substitution e170N | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit, ... | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
7QVI
 
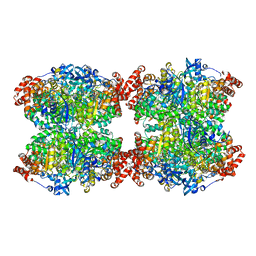 | | Fiber-forming RubisCO derived from ancestral sequence reconstruction and rational engineering | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit | | Authors: | Schulz, L, Zarzycki, J, Prinz, S, Schuller, J.M, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2022-01-21 | | Release date: | 2022-10-12 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
7QSW
 
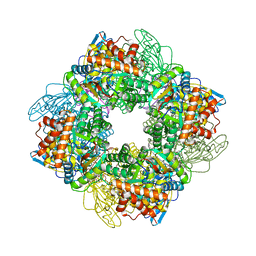 | | L8S8-complex forming RubisCO derived from ancestral sequence reconstruction of the last common ancestor of SSU-bearing Form I RubisCOs | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit, ... | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
7QSV
 
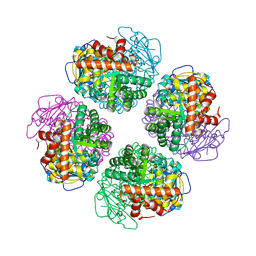 | | L8-complex forming RubisCO derived from ancestral sequence reconstruction of the last common ancestor of Form I'' and Form I RubisCOs | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
7QSY
 
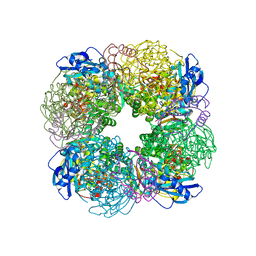 | | Non-obligately L8S8-complex forming RubisCO derived from ancestral sequence reconstruction and rational engineering in L8S8 complex | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit, ... | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
8AEX
 
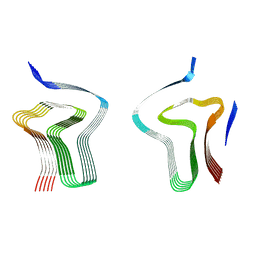 | | Lipidic alpha-synuclein fibril - polymorph L3A | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-07-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
7QSZ
 
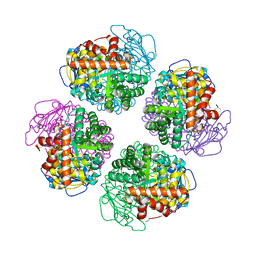 | | Non-obligately L8S8-complex forming RubisCO derived from ancestral sequence reconstruction and rational engineering in L8 complex with substitution e170N | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, RubisCO large subunit | | Authors: | Zarzycki, J, Schulz, L, Erb, T.J, Hochberg, G.K.A. | | Deposit date: | 2022-01-14 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Evolution of increased complexity and specificity at the dawn of form I Rubiscos.
Science, 378, 2022
|
|
8ADV
 
 | | Lipidic alpha-synuclein fibril - polymorph L1B | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-07-11 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.98 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
8ADS
 
 | | Lipidic alpha-synuclein fibril - polymorph L2B | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-07-11 | | Release date: | 2022-10-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
8A2U
 
 | | Cryo-EM structure of F-actin in the Ca2+-ADP-BeF3- nucleotide state. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Oosterheert, W, Klink, B.U, Belyy, A, Pospich, S, Raunser, S. | | Deposit date: | 2022-06-06 | | Release date: | 2022-08-10 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.21 Å) | | Cite: | Structural basis of actin filament assembly and aging.
Nature, 611, 2022
|
|
8A2T
 
 | | Cryo-EM structure of F-actin in the Mg2+-ADP nucleotide state. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Oosterheert, W, Klink, B.U, Belyy, A, Pospich, S, Raunser, S. | | Deposit date: | 2022-06-06 | | Release date: | 2022-08-10 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.24 Å) | | Cite: | Structural basis of actin filament assembly and aging.
Nature, 611, 2022
|
|
8A2Z
 
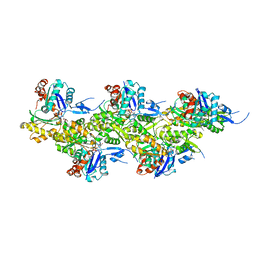 | | Cryo-EM structure of F-actin in the Ca2+-ADP nucleotide state. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Oosterheert, W, Klink, B.U, Belyy, A, Pospich, S, Raunser, S. | | Deposit date: | 2022-06-06 | | Release date: | 2022-08-10 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Structural basis of actin filament assembly and aging.
Nature, 611, 2022
|
|
8A4C
 
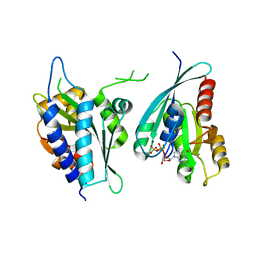 | | Structure of human Rep15:Rab3B complex. | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Rab15 effector protein, ... | | Authors: | Rai, A, Vetter, I.R, Goody, R.S. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Rep15 interacts with several Rab GTPases and has a distinct fold for a Rab effector.
Nat Commun, 13, 2022
|
|
8A2Y
 
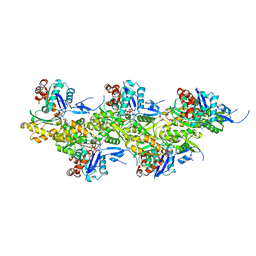 | | Cryo-EM structure of F-actin in the Ca2+-ADP-Pi nucleotide state. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Oosterheert, W, Klink, B.U, Belyy, A, Pospich, S, Raunser, S. | | Deposit date: | 2022-06-06 | | Release date: | 2022-08-10 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Structural basis of actin filament assembly and aging.
Nature, 611, 2022
|
|
8A2S
 
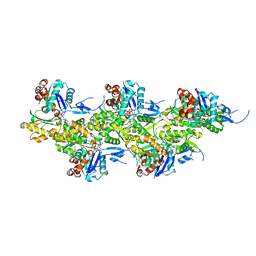 | | Cryo-EM structure of F-actin in the Mg2+-ADP-Pi nucleotide state. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Oosterheert, W, Klink, B.U, Belyy, A, Pospich, S, Raunser, S. | | Deposit date: | 2022-06-06 | | Release date: | 2022-08-10 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.22 Å) | | Cite: | Structural basis of actin filament assembly and aging.
Nature, 611, 2022
|
|
8A4B
 
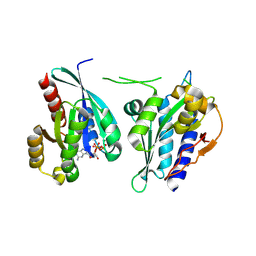 | | Structure of human Rep15:Rab3B_Q81L complex. | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Rab15 effector protein, ... | | Authors: | Rai, A, Vetter, I.R, Goody, R.S. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Rep15 interacts with several Rab GTPases and has a distinct fold for a Rab effector.
Nat Commun, 13, 2022
|
|
8A2R
 
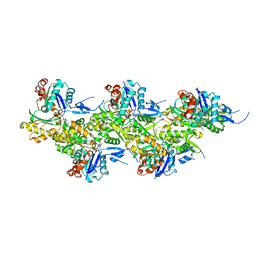 | | Cryo-EM structure of F-actin in the Mg2+-ADP-BeF3- nucleotide state. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Oosterheert, W, Klink, B.U, Belyy, A, Pospich, S, Raunser, S. | | Deposit date: | 2022-06-06 | | Release date: | 2022-08-10 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.17 Å) | | Cite: | Structural basis of actin filament assembly and aging.
Nature, 611, 2022
|
|
8A4A
 
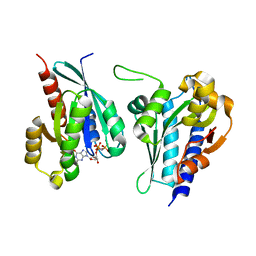 | | Structure of human Rep15 in complex with bovine Rab3C. | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Rab15 effector protein, ... | | Authors: | Rai, A, Vetter, I.R, Goody, R.S. | | Deposit date: | 2022-06-10 | | Release date: | 2022-08-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Rep15 interacts with several Rab GTPases and has a distinct fold for a Rab effector.
Nat Commun, 13, 2022
|
|
8A4L
 
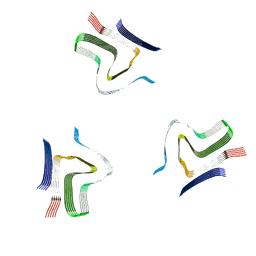 | | Lipidic alpha-synuclein fibril - polymorph L2A | | Descriptor: | Alpha-synuclein | | Authors: | Frieg, B, Antonschmidt, L, Dienemann, C, Geraets, J.A, Najbauer, E.E, Matthes, D, de Groot, B.L, Andreas, L.B, Becker, S, Griesinger, C, Schroeder, G.F. | | Deposit date: | 2022-06-12 | | Release date: | 2022-08-17 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | The 3D structure of lipidic fibrils of alpha-synuclein.
Nat Commun, 13, 2022
|
|
8APR
 
 | |
7ZDV
 
 | |
7ZDA
 
 | |
7ZBZ
 
 | |
