6BXU
 
 | |
6EET
 
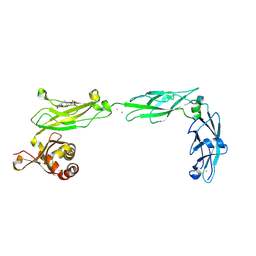 | | Crystal structure of mouse Protocadherin-15 EC9-MAD12 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Narui, Y, Sotomayor, M. | | Deposit date: | 2018-08-15 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structural determinants of protocadherin-15 mechanics and function in hearing and balance perception.
Proc.Natl.Acad.Sci.USA, 2020
|
|
5W1D
 
 | |
6E8F
 
 | |
6EB5
 
 | |
1L3W
 
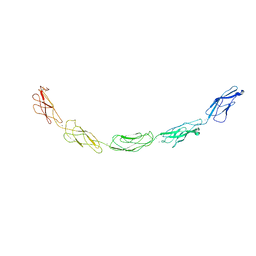 | | C-cadherin Ectodomain | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Boggon, T.J, Murray, J, Chappuis-Flament, S, Wong, E, Gumbiner, B.M, Shapiro, L. | | Deposit date: | 2002-03-01 | | Release date: | 2002-04-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.08 Å) | | Cite: | C-cadherin ectodomain structure and implications for cell adhesion mechanisms
Science, 296, 2002
|
|
1Q6A
 
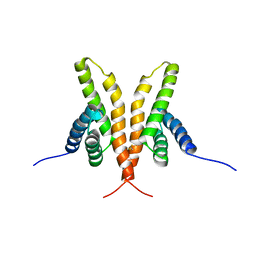 | | Solution Structure of the C-terminal Domain of Thermosynechococcus elongatus KaiA (ThKaiA180C); Averaged Minimized Structure | | Descriptor: | Circadian clock protein KaiA homolog | | Authors: | Vakonakis, I, Sun, J, Holzenburg, A, Golden, S.S, LiWang, A.C. | | Deposit date: | 2003-08-13 | | Release date: | 2003-08-19 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: Implications for KaiA-KaiC interaction
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1Q6B
 
 | | Solution Structure of the C-terminal Domain of Thermosynechococcus elongatus KaiA (ThKaiA180C); Ensemble of 25 Structures | | Descriptor: | Circadian clock protein KaiA homolog | | Authors: | Vakonakis, I, Sun, J, Golden, S.S, Holzenburg, A, LiWang, A.C. | | Deposit date: | 2003-08-13 | | Release date: | 2003-08-19 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the KaiC-interacting C-terminal domain of KaiA, a circadian clock protein: implications for KaiA-KaiC interaction
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
3K6F
 
 | |
3K6I
 
 | |
6PSF
 
 | | Rhinovirus C15 complexed with domains I and II of receptor CDHR3 | | Descriptor: | Cadherin-related family member 3, Capsid protein VP1, Capsid protein VP2, ... | | Authors: | Sun, Y, Watters, K, Klose, T, Palmenberg, A.C. | | Deposit date: | 2019-07-12 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of rhinovirus C15a bound to its cadherin-related protein 3 receptor.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5F53
 
 | |
2V37
 
 | | Solution structure of the N-terminal extracellular domain of human T- cadherin | | Descriptor: | CADHERIN-13 | | Authors: | Dames, S.A, Bang, E.J, Ahrens, T, Haeussinger, D, Grzesiek, S. | | Deposit date: | 2007-06-13 | | Release date: | 2008-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Insights into the low adhesive capacity of human T-cadherin from the NMR structure of Its N-terminal extracellular domain.
J. Biol. Chem., 283, 2008
|
|
6PPO
 
 | | Rhinovirus C15 complexed with domain I of receptor CDHR3 | | Descriptor: | CALCIUM ION, Cadherin-related family member 3, Capsid protein VP1, ... | | Authors: | Sun, Y, Watters, K, Klose, T, Palmenberg, A.C. | | Deposit date: | 2019-07-08 | | Release date: | 2020-03-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of rhinovirus C15a bound to its cadherin-related protein 3 receptor.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6MEQ
 
 | | PcdhgB3 EC1-4 in 50 mM HEPES | | Descriptor: | CALCIUM ION, Protocadherin gamma-B3 | | Authors: | Nicoludis, J.M, Gaudet, R. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Interaction specificity of clustered protocadherins inferred from sequence covariation and structural analysis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6MER
 
 | | PcdhgB3 EC1-4 in 50 mM HEPES | | Descriptor: | CALCIUM ION, Protocadherin gamma-B3 | | Authors: | Nicoludis, J.M, Gaudet, R. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Interaction specificity of clustered protocadherins inferred from sequence covariation and structural analysis.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1U9I
 
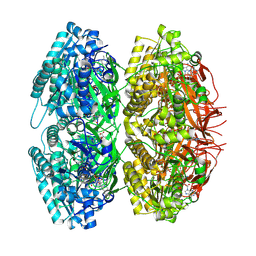 | | Crystal Structure of Circadian Clock Protein KaiC with Phosphorylation Sites | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, KaiC, MAGNESIUM ION | | Authors: | Xu, Y, Mori, T, Pattanayek, R, Pattanayek, S, Egli, M, Johnson, C.H. | | Deposit date: | 2004-08-09 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of key phosphorylation sites in the circadian clock protein KaiC by crystallographic and mutagenetic analyses
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
1CDP
 
 | |
1NZ4
 
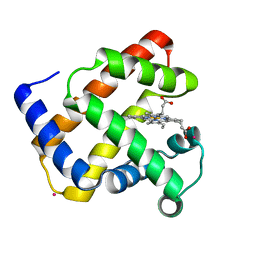 | | The horse heart myoglobin variant K45E/K63E complexed with Cadmium | | Descriptor: | CADMIUM ION, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hunter, C.L, Maurus, R, Mauk, M.R, Lee, H, Raven, E.L, Tong, H, Nguyen, N, Smith, S, Brayer, G.D, Mauk, A.G. | | Deposit date: | 2003-02-15 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Introduction and characterization of a functionally linked metal ion binding site at
the exposed heme edge of myoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
2QVK
 
 | |
2QVM
 
 | | The second Ca2+-binding domain of the Na+-Ca2+ exchanger is essential for regulation: crystal structures and mutational analysis | | Descriptor: | CALCIUM ION, Sodium/calcium exchanger 1 | | Authors: | Chaptal, V, Mercado Besserer, G, Abramson, J, Cascio, D. | | Deposit date: | 2007-08-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | The second Ca2+-binding domain of the Na+ Ca2+ exchanger is essential for regulation: crystal structures and mutational analysis
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3K5S
 
 | |
5V5X
 
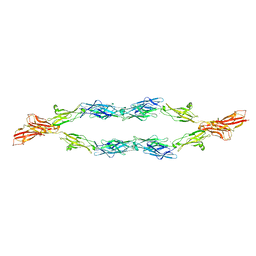 | | Protocadherin gammaB7 EC3-6 cis-dimer structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Goodman, K.M, Mannepalli, S, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2017-03-15 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Protocadherin cis-dimer architecture and recognition unit diversity.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8AP5
 
 | |
1OP4
 
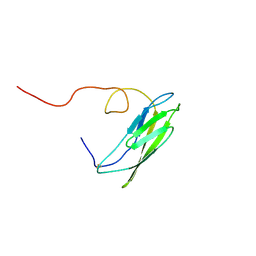 | | Solution Structure of Neural Cadherin Prodomain | | Descriptor: | Neural-cadherin | | Authors: | Koch, A.W, Farooq, A, Shan, W, Zeng, L, Colman, D.R, Zhou, M.-M. | | Deposit date: | 2003-03-04 | | Release date: | 2004-03-16 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Structure of the Neural (N-) Cadherin Prodomain Reveals a Cadherin Extracellular Domain-like Fold without Adhesive Characteristics
Structure, 12, 2004
|
|
