1RBF
 
 | |
1RDB
 
 | |
1RBI
 
 | |
2BPP
 
 | |
1RIL
 
 | | CRYSTAL STRUCTURE OF RIBONUCLEASE H FROM THERMUS THERMOPHILUS HB8 REFINED AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | RIBONUCLEASE H | | Authors: | Ishikawa, K, Okumura, M, Katayanagi, K, Kimura, S, Kanaya, S, Nakamura, H, Morikawa, K. | | Deposit date: | 1993-01-14 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ribonuclease H from Thermus thermophilus HB8 refined at 2.8 A resolution.
J.Mol.Biol., 230, 1993
|
|
1RAS
 
 | | CRYSTAL STRUCTURE OF A FLUORESCENT DERIVATIVE OF RNASE A | | Descriptor: | 5-(1-SULFONAPHTHYL)-ACETYLAMINO-ETHYLAMINE, RIBONUCLEASE A | | Authors: | Baudet-Nessler, S, Jullien, M, Crosio, M.-P, Janin, J. | | Deposit date: | 1993-03-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of a fluorescent derivative of RNase A.
Biochemistry, 32, 1993
|
|
1RAR
 
 | | CRYSTAL STRUCTURE OF A FLUORESCENT DERIVATIVE OF RNASE A | | Descriptor: | 5-(1-SULFONAPHTHYL)-ACETYLAMINO-ETHYLAMINE, CHLORIDE ION, RIBONUCLEASE A | | Authors: | Baudet-Nessler, S, Jullien, M, Crosio, M.-P, Janin, J. | | Deposit date: | 1993-03-02 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a fluorescent derivative of RNase A.
Biochemistry, 32, 1993
|
|
2TIR
 
 | | CRYSTAL STRUCTURE ANALYSIS OF A MUTANT ESCHERICHIA COLI THIOREDOXIN IN WHICH LYSINE 36 IS REPLACED BY GLUTAMIC ACID | | Descriptor: | COPPER (II) ION, THIOREDOXIN | | Authors: | Nikkola, M, Gleason, F.K, Fuchs, J.A, Eklund, H. | | Deposit date: | 1993-01-10 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure analysis of a mutant Escherichia coli thioredoxin in which lysine 36 is replaced by glutamic acid.
Biochemistry, 32, 1993
|
|
1HOM
 
 | | DETERMINATION OF THE THREE-DIMENSIONAL STRUCTURE OF THE ANTENNAPEDIA HOMEODOMAIN FROM DROSOPHILA IN SOLUTION BY 1H NUCLEAR MAGNETIC RESONANCE SPECTROSCOPY | | Descriptor: | ANTENNAPEDIA PROTEIN | | Authors: | Qian, Y.-Q, Billeter, M, Otting, G, Muller, M, Gehring, W.J, Wuthrich, K. | | Deposit date: | 1991-10-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Determination of the three-dimensional structure of the Antennapedia homeodomain from Drosophila in solution by 1H nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 214, 1990
|
|
2RN2
 
 | | STRUCTURAL DETAILS OF RIBONUCLEASE H FROM ESCHERICHIA COLI AS REFINED TO AN ATOMIC RESOLUTION | | Descriptor: | RIBONUCLEASE H | | Authors: | Katayanagi, K, Miyagawa, M, Matsushima, M, Ishikawa, M, Kanaya, S, Nakamura, H, Ikehara, M, Matsuzaki, T, Morikawa, K. | | Deposit date: | 1992-04-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural details of ribonuclease H from Escherichia coli as refined to an atomic resolution.
J.Mol.Biol., 223, 1992
|
|
2PK4
 
 | |
1IXA
 
 | | THE THREE-DIMENSIONAL STRUCTURE OF THE FIRST EGF-LIKE MODULE OF HUMAN FACTOR IX: COMPARISON WITH EGF AND TGF-A | | Descriptor: | EGF-LIKE MODULE OF HUMAN FACTOR IX | | Authors: | Baron, M, Norman, D.G, Harvey, T.S, Hanford, P.A, Mayhew, M, Tse, A.G.D, Brownlee, G.G, Campbell, I.D.C. | | Deposit date: | 1991-11-14 | | Release date: | 1993-10-31 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the first EGF-like module of human factor IX: comparison with EGF and TGF-alpha.
Protein Sci., 1, 1992
|
|
1AAL
 
 | |
1RBD
 
 | |
1RCM
 
 | |
6LYT
 
 | |
1RLC
 
 | | CRYSTAL STRUCTURE OF THE UNACTIVATED RIBULOSE 1, 5-BISPHOSPHATE CARBOXYLASE(SLASH)OXYGENASE COMPLEXED WITH A TRANSITION STATE ANALOG, 2-CARBOXY-D-ARABINITOL 1,5-BISPHOSPHATE | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (LARGE CHAIN), RIBULOSE 1,5 BISPHOSPHATE CARBOXYLASE/OXYGENASE (SMALL CHAIN) | | Authors: | Zhang, K.Y.J, Cascio, D, Eisenberg, D. | | Deposit date: | 1993-08-04 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the unactivated ribulose 1,5-bisphosphate carboxylase/oxygenase complexed with a transition state analog, 2-carboxy-D-arabinitol 1,5-bisphosphate.
Protein Sci., 3, 1994
|
|
1RDA
 
 | |
1RBH
 
 | |
1ATR
 
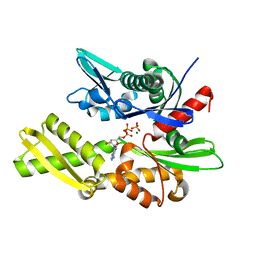 | |
1GMC
 
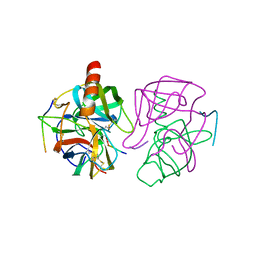 | | THE X-RAY CRYSTAL STRUCTURE OF THE TETRAHEDRAL INTERMEDIATE OF GAMMA-CHYMOTRYPSIN IN HEXANE | | Descriptor: | GAMMA-CHYMOTRYPSIN A, PRO GLY ALA TYR PEPTIDE | | Authors: | Yennawar, N.H, Yennawar, H.P, Banerjee, S, Farber, G.K. | | Deposit date: | 1993-08-20 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystal structure of gamma-chymotrypsin in hexane.
Biochemistry, 33, 1994
|
|
4GST
 
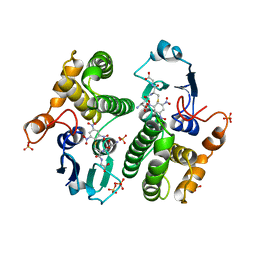 | | REACTION COORDINATE MOTION IN AN SNAR REACTION CATALYZED BY GLUTATHIONE TRANSFERASE | | Descriptor: | 1-(S-GLUTATHIONYL)-2,4,6-TRINITROCYCLOHEXA-2,5-DIENE, GLUTATHIONE S-TRANSFERASE, SULFATE ION | | Authors: | Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1993-07-20 | | Release date: | 1993-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Snapshots along the reaction coordinate of an SNAr reaction catalyzed by glutathione transferase.
Biochemistry, 32, 1993
|
|
1GMD
 
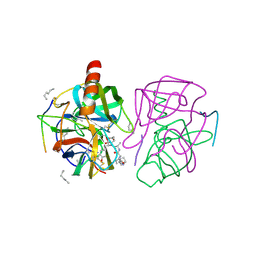 | | X-ray crystal structure of gamma-chymotrypsin in hexane | | Descriptor: | GAMMA-CHYMOTRYPSIN A, HEXANE, PRO GLY ALA TYR ASP PEPTIDE | | Authors: | Yennawar, N.H, Yennawar, H.P, Banerjee, S, Farber, G.K. | | Deposit date: | 1993-08-20 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray crystal structure of gamma-chymotrypsin in hexane.
Biochemistry, 33, 1994
|
|
1GHL
 
 | |
1KDU
 
 | | SEQUENTIAL 1H NMR ASSIGNMENTS AND SECONDARY STRUCTURE OF THE KRINGLE DOMAIN FROM UROKINASE | | Descriptor: | PLASMINOGEN ACTIVATOR | | Authors: | Li, X, Bokman, A.M, Llinas, M, Smith, R.A.G, Dobson, C.M. | | Deposit date: | 1993-07-15 | | Release date: | 1993-10-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the kringle domain from urokinase-type plasminogen activator.
J.Mol.Biol., 235, 1994
|
|
