4NTC
 
 | | Crystal structure of GliT | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GliT | | Authors: | Scharf, D.H, Groll, M, Habel, A, Heinekamp, T, Hertweck, C, Brakhage, A.A, Huber, E.M. | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flavoenzyme-Catalyzed Formation of Disulfide Bonds in Natural Products
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NTE
 
 | | Crystal structure of DepH | | Descriptor: | CHLORIDE ION, DepH, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Scharf, D.H, Groll, M, Habel, A, Heinekamp, T, Hertweck, C, Brakhage, A.A, Huber, E.M. | | Deposit date: | 2013-12-02 | | Release date: | 2014-03-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flavoenzyme-Catalyzed Formation of Disulfide Bonds in Natural Products
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2ASM
 
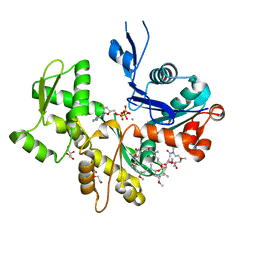 | | Structure of Rabbit Actin In Complex With Reidispongiolide A | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Allingham, J.S, Zampella, A, D'Auria, M.V, Rayment, I. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of microfilament destabilizing toxins bound to actin provide insight into toxin design and activity
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1IOL
 
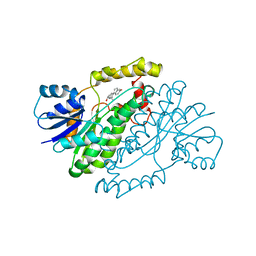 | | ESTROGENIC 17-BETA HYDROXYSTEROID DEHYDROGENASE COMPLEXED 17-BETA-ESTRADIOL | | Descriptor: | ESTRADIOL, ESTROGENIC 17-BETA HYDROXYSTEROID DEHYDROGENASE | | Authors: | Azzi, A, Rehse, P.H, Zhu, D.-W, Campbell, R.L, Labrie, F, Lin, S.-X. | | Deposit date: | 1996-06-20 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of human estrogenic 17 beta-hydroxysteroid dehydrogenase complexed with 17 beta-estradiol.
Nat.Struct.Biol., 3, 1996
|
|
3MLQ
 
 | |
5LFA
 
 | | Crystal structure of iron-sulfur cluster containing bacterial (6-4) photolyase PhrB - Y424F mutant with impaired DNA repair activity | | Descriptor: | (6-4) photolyase, 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kwiatkowski, D, Zhang, F, Krauss, N, Lamparter, T, Scheerer, P. | | Deposit date: | 2016-06-30 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structures of Bacterial (6-4) Photolyase Mutants with Impaired DNA Repair Activity.
Photochem. Photobiol., 93, 2017
|
|
5X2I
 
 | |
4PZY
 
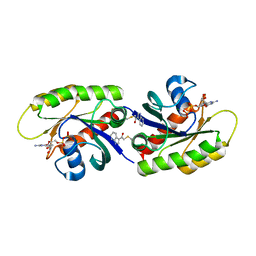 | | Second-site screening of K-Ras in the presence of covalently attached first-site ligands | | Descriptor: | 2-chloro-1-(1H-indol-3-yl)ethanone, GUANOSINE-5'-DIPHOSPHATE, K-Ras, ... | | Authors: | Sun, Q, Phan, J, Friberg, A, Camper, D.V, Olejniczak, E.T, Fesik, S.W. | | Deposit date: | 2014-03-31 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | A method for the second-site screening of K-Ras in the presence of a covalently attached first-site ligand.
J.Biomol.Nmr, 60, 2014
|
|
2ASP
 
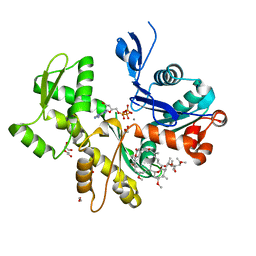 | | Structure of Rabbit Actin In Complex With Reidispongiolide C | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, Actin, ... | | Authors: | Allingham, J.S, Zampella, A, D'Auria, M.V, Rayment, I. | | Deposit date: | 2005-08-23 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structures of microfilament destabilizing toxins bound to actin provide insight into toxin design and activity
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
2AUX
 
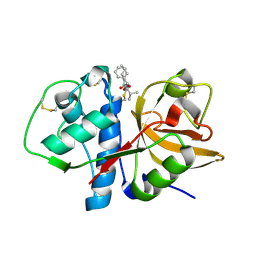 | | Cathepsin K complexed with a semicarbazone inhibitor | | Descriptor: | (1R)-2-METHYL-1-(PHENYLMETHYL)PROPYL[(1S)-1-FORMYLPENTYL]CARBAMATE, Cathepsin K | | Authors: | Adkison, K.K, Barrett, D.G, Deaton, D.N, Gampe, R.T, Hassell, A.M, Long, S.T, McFadyen, R.B, Miller, A.B, Miller, L.R, Shewchuk, L.M. | | Deposit date: | 2005-08-29 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Semicarbazone-based inhibitors of cathepsin K, are they prodrugs for aldehyde inhibitors?
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2C2S
 
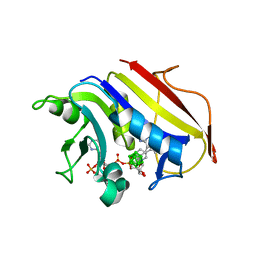 | | Human Dihydrofolate Reductase Complexed With NADPH and 2,4-Diamino-5-(1-o-carboranylmethyl)-6-methylpyrimidine, A novel boron containing, nonclassical Antifolate | | Descriptor: | 2,4-DIAMINO-5-(1-O-CARBORANYLMETHYL)-6-METHYLPYRIMIDINE, DIHYDROFOLATE REDUCTASE, GLYCEROL, ... | | Authors: | Leung, A.K.W, Reynolds, R.C, Riordan, J.M, Borhani, D.W. | | Deposit date: | 2005-09-29 | | Release date: | 2007-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Novel Boron-Containing, Nonclassical Antifolates: Synthesis and Preliminary Biological and Structural Evaluation.
J.Med.Chem., 50, 2007
|
|
2C4C
 
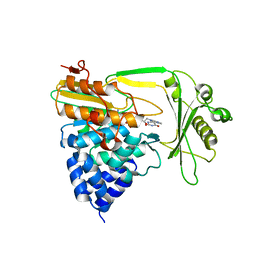 | | Crystal structure of the NADPH-treated monooxygenase domain of MICAL | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, NEDD9-INTERACTING PROTEIN WITH CALPONIN HOMOLOGY AND LIM DOMAINS | | Authors: | Siebold, C, Berrow, N, Walter, T.S, Harlos, K, Owens, R.J, Terman, J.R, Stuart, D.I, Kolodkin, A.L, Pasterkamp, R.J, Jones, E.Y. | | Deposit date: | 2005-10-18 | | Release date: | 2005-10-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | High-Resolution Structure of the Catalytic Region of Mical (Molecule Interacting with Casl), a Multidomain Flavoenzyme-Signaling Molecule.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
4Q01
 
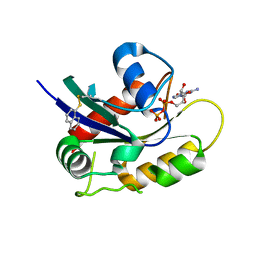 | | Second-site screening of K-Ras in the presence of covalently attached first-site ligands | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, K-Ras, MAGNESIUM ION, ... | | Authors: | Sun, Q, Phan, J, Friberg, A, Camper, D.V, Olejniczak, E.T, Fesik, S.W. | | Deposit date: | 2014-03-31 | | Release date: | 2014-09-10 | | Method: | X-RAY DIFFRACTION (1.291 Å) | | Cite: | A method for the second-site screening of K-Ras in the presence of a covalently attached first-site ligand.
J.Biomol.Nmr, 60, 2014
|
|
5JYC
 
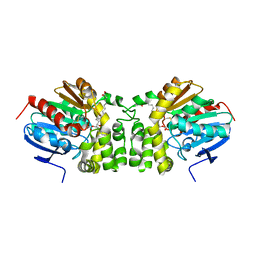 | | Crystal structure of the E153Q mutant of the CFTR inhibitory factor Cif containing the adducted 14,15-EET hydrolysis intermediate | | Descriptor: | (5~{Z},11~{Z},14~{R},15~{R})-14,15-bis(oxidanyl)icosa-5,8,11-trienoic acid, CFTR inhibitory factor | | Authors: | Hvorecny, K.L, Madden, D.R. | | Deposit date: | 2016-05-13 | | Release date: | 2017-01-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Pseudomonas aeruginosa sabotages the generation of host proresolving lipid mediators.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7C0P
 
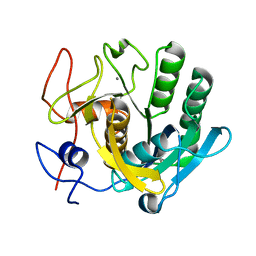 | |
5TVK
 
 | |
5HMN
 
 | | Crystal structure of an aminoglycoside acetyltransferase HMB0005 from an uncultured soil metagenomic sample, unknown active site density modeled as polyethylene glycol | | Descriptor: | AAC3-I, COENZYME A, TETRAETHYLENE GLYCOL | | Authors: | Xu, Z, Stogios, P.J, Wawrzak, Z, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-16 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.018 Å) | | Cite: | Crystal structure of an aminoglycoside acetyltransferase HMB0005 from an uncultured soil metagenomic sample, unknown active site density modeled as polyethylene glycol
To Be Published
|
|
1I4A
 
 | | CRYSTAL STRUCTURE OF PHOSPHORYLATION-MIMICKING MUTANT T6D OF ANNEXIN IV | | Descriptor: | ANNEXIN IV, CALCIUM ION, SULFATE ION | | Authors: | Kaetzel, M.A, Mo, Y.D, Mealy, T.R, Campos, B, Bergsma-Schutter, W, Brisson, A, Dedman, J.R, Seaton, B.A. | | Deposit date: | 2001-02-20 | | Release date: | 2001-04-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylation mutants elucidate the mechanism of annexin IV-mediated membrane aggregation.
Biochemistry, 40, 2001
|
|
2ZW2
 
 | | Crystal Structure of Formylglycinamide Ribonucleotide Amidotransferase III from SULFOLOBUS TOKODAII (STPURS) | | Descriptor: | GLYCEROL, Putative uncharacterized protein STS178 | | Authors: | Suzuki, S, Tamura, S, Okada, K, Baba, S, Kumasaka, T, Nakagawa, N, Masui, R, Kuramitsu, S, Sampei, G, Kawai, G. | | Deposit date: | 2008-11-27 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Formylglycinamide Ribonucleotide Amidotransferase III from SULFOLOBUS TOKODAII (STPURS)
To be Published
|
|
5JCG
 
 | |
2AUZ
 
 | | Cathepsin K complexed with a semicarbazone inhibitor | | Descriptor: | 1-(PHENYLMETHYL)CYCLOPENTYL[(1S)-1-FORMYLPENTYL]CARBAMATE, Cathepsin K, SULFATE ION | | Authors: | Adkison, K.K, Barrett, D.G, Deaton, D.N, Gampe, R.T, Hassell, A.M, Long, S.T, McFadyen, R.B, Miller, A.B, Miller, L.R, Shewchuk, L.M. | | Deposit date: | 2005-08-29 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Semicarbazone-based inhibitors of cathepsin K, are they prodrugs for aldehyde inhibitors?
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1G6C
 
 | | THIAMIN PHOSPHATE SYNTHASE | | Descriptor: | 2-TRIFLUOROMETHYL-5-METHYLENE-5H-PYRIMIDIN-4-YLIDENEAMINE, 4-METHYL-5-HYDROXYETHYLTHIAZOLE PHOSPHATE, MAGNESIUM ION, ... | | Authors: | Peapus, D.H, Chiu, H.-J, Campobasso, N, Reddick, J.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-11-03 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural characterization of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes of thiamin phosphate synthase.
Biochemistry, 40, 2001
|
|
1GGU
 
 | | HUMAN FACTOR XIII WITH CALCIUM BOUND IN THE ION SITE | | Descriptor: | CALCIUM ION, PROTEIN (COAGULATION FACTOR XIII) | | Authors: | Fox, B.A, Yee, V.C, Pederson, L.C, Trong, I.L, Bishop, P.D, Stenkamp, R.E, Teller, D.C. | | Deposit date: | 1998-07-22 | | Release date: | 1999-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Identification of the calcium binding site and a novel ytterbium site in blood coagulation factor XIII by x-ray crystallography.
J.Biol.Chem., 274, 1999
|
|
5FJO
 
 | |
1G4E
 
 | | THIAMIN PHOSPHATE SYNTHASE | | Descriptor: | THIAMIN PHOSPHATE SYNTHASE | | Authors: | Peapus, D.H, Chiu, H.-J, Campobasso, N, Reddick, J.J, Begley, T.P, Ealick, S.E. | | Deposit date: | 2000-10-26 | | Release date: | 2001-09-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of the enzyme-substrate, enzyme-intermediate, and enzyme-product complexes of thiamin phosphate synthase.
Biochemistry, 40, 2001
|
|
