2QYV
 
 | |
4K61
 
 | |
2R1I
 
 | |
2QCK
 
 | |
4KQ7
 
 | |
2QZC
 
 | |
6AI1
 
 | |
2QTP
 
 | |
2R4I
 
 | |
4PS6
 
 | |
2Q7B
 
 | |
6AL3
 
 | | Lys49 PLA2 BPII derived from the venom of Protobothrops flavoviridis. | | Descriptor: | Basic phospholipase A2 BP-II, SULFATE ION | | Authors: | Matsui, T, Kamata, S, Suzuki, A, Oda-Ueda, N, Ogawa, T, Tanaka, Y. | | Deposit date: | 2018-09-05 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | SDS-induced oligomerization of Lys49-phospholipase A2from snake venom.
Sci Rep, 9, 2019
|
|
4Q34
 
 | |
6AL2
 
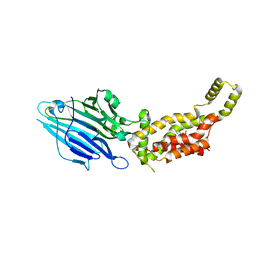 | | Crystal structure of E. coli YidC at 2.8 A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Membrane protein insertase YidC | | Authors: | Tanaka, Y, Tsukazaki, T, Izumioka, A, Hamid, A.A, Fujii, A. | | Deposit date: | 2018-09-05 | | Release date: | 2018-11-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8-angstrom crystal structure of Escherichia coli YidC revealing all core regions, including flexible C2 loop.
Biochem. Biophys. Res. Commun., 505, 2018
|
|
4IVE
 
 | |
2QDR
 
 | |
2QEA
 
 | |
2QEU
 
 | |
4Q0Y
 
 | |
4KQT
 
 | |
4Q6K
 
 | |
2Q7X
 
 | |
2Q78
 
 | |
2PY6
 
 | |
2Q9K
 
 | |
