4C6I
 
 | | Crystal structure of the dihydroorotase domain of human CAD bound to substrate at pH 7.0 | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD PROTEIN, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
4C6D
 
 | | Crystal structure of the dihydroorotase domain of human CAD bound to substrate at pH 6.0 | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD PROTEIN, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.298 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
4C6L
 
 | | Crystal structure of the dihydroorotase domain of human CAD bound to the inhibitor fluoroorotate at pH 6.0 | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD PROTEIN, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
4C6J
 
 | | Crystal structure of the dihydroorotase domain of human CAD bound to substrate at pH 7.5 | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD PROTEIN, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Lallous, N, Grande-Garcia, A. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.299 Å) | | Cite: | Structure, Functional Characterization and Evolution of the Dihydroorotase Domain of Human Cad.
Structure, 22, 2014
|
|
6HG3
 
 | | Hybrid dihydroorotase domain of human CAD with E. coli flexible loop, bound to dihydroorotate | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD protein, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Del Cano-Ochoa, F. | | Deposit date: | 2018-08-22 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Characterization of the catalytic flexible loop in the dihydroorotase domain of the human multi-enzymatic protein CAD.
J. Biol. Chem., 293, 2018
|
|
6HG2
 
 | | Hybrid dihydroorotase domain of human CAD with E. coli flexible loop, bound to fluoroorotate | | Descriptor: | 5-FLUORO-2,6-DIOXO-1,2,3,6-TETRAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, CAD protein, FORMIC ACID, ... | | Authors: | Ramon-Maiques, S, Del Cano-Ochoa, F. | | Deposit date: | 2018-08-22 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Characterization of the catalytic flexible loop in the dihydroorotase domain of the human multi-enzymatic protein CAD.
J. Biol. Chem., 293, 2018
|
|
5VKT
 
 | | Cinnamyl alcohol dehydrogenases (SbCAD4) from Sorghum bicolor (L.) Moench | | Descriptor: | ACETATE ION, Cinnamyl alcohol dehydrogenases (SbCAD4), D-MALATE, ... | | Authors: | Walker, A.M, Jun, S.Y, Kang, C. | | Deposit date: | 2017-04-22 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.827 Å) | | Cite: | The Enzyme Activity and Substrate Specificity of Two Major Cinnamyl Alcohol Dehydrogenases in Sorghum (Sorghum bicolor), SbCAD2 and SbCAD4.
Plant Physiol., 174, 2017
|
|
5LG8
 
 | | Human L-type ferritin iron loaded for 60 minutes | | Descriptor: | CADMIUM ION, FE (III) ION, Ferritin light chain, ... | | Authors: | Pozzi, C, Di Pisa, F, Mangani, S. | | Deposit date: | 2016-07-06 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Chemistry at the protein-mineral interface in L-ferritin assists the assembly of a functional ( mu (3)-oxo)Tris[( mu (2)-peroxo)] triiron(III) cluster.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5LG2
 
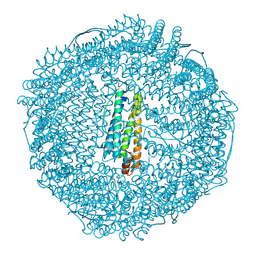 | | Horse L type ferritin iron loaded for 60 minutes | | Descriptor: | CADMIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Pozzi, C, Di Pisa, F, Mangani, S. | | Deposit date: | 2016-07-05 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Chemistry at the protein-mineral interface in L-ferritin assists the assembly of a functional ( mu (3)-oxo)Tris[( mu (2)-peroxo)] triiron(III) cluster.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5CPV
 
 | |
3Q2B
 
 | | Crystal Structure of an Actin Depolymerizing Factor | | Descriptor: | BETA-MERCAPTOETHANOL, Cofilin/actin-depolymerizing factor homolog 1, D(-)-TARTARIC ACID | | Authors: | Wong, W, Clarke, O.B, Gulbis, J.M, Baum, J. | | Deposit date: | 2010-12-19 | | Release date: | 2011-06-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Minimal requirements for actin filament disassembly revealed by structural analysis of malaria parasite actin-depolymerizing factor 1
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2KAD
 
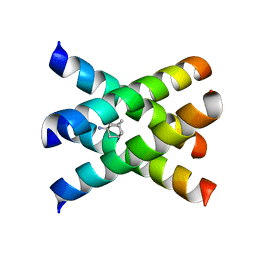 | | Magic-Angle-Spinning Solid-State NMR Structure of Influenza A M2 Transmembrane Domain | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, Transmembrane peptide of Matrix protein 2 | | Authors: | Hong, M, Cady, S.D, Mishanina, T.V. | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure of amantadine-bound M2 transmembrane peptide of influenza A in lipid bilayers from magic-angle-spinning solid-state NMR: the role of Ser31 in amantadine binding.
J.Mol.Biol., 385, 2009
|
|
1C1J
 
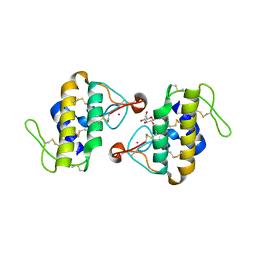 | | STRUCTURE OF CADMIUM-SUBSTITUTED PHOSPHOLIPASE A2 FROM AGKISTRONDON HALYS PALLAS AT 2.8 ANGSTROMS RESOLUTION | | Descriptor: | BASIC PHOSPHOLIPASE A2, CADMIUM ION, octyl beta-D-glucopyranoside | | Authors: | Zhang, H.-l, Zhang, Y.-q, Song, S.-y, Zhou, Y, Lin, Z.-j. | | Deposit date: | 1999-07-22 | | Release date: | 2002-07-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of Cadmium-substituted Phospholipase A2 from Agkistrodon halys
Pallas at 2.8 Angstroms Resolution
Protein Pept.Lett., 6, 1999
|
|
5T5X
 
 | |
4U7U
 
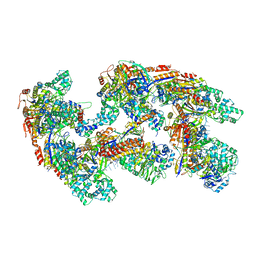 | | Crystal structure of RNA-guided immune Cascade complex from E.coli | | Descriptor: | CRISPR system Cascade subunit CasA, CRISPR system Cascade subunit CasB, CRISPR system Cascade subunit CasC, ... | | Authors: | Zhao, H, Sheng, G, Wang, J, Wang, M, Bunkoczi, G, Gong, W, Wei, Z, Wang, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-08-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli
Nature, 515, 2014
|
|
6JEF
 
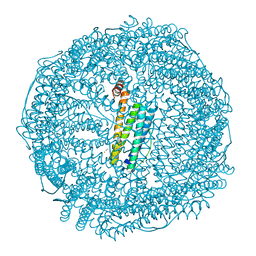 | | Crystal structure of apo-R168C/L169C-Fr | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferritin light chain, ... | | Authors: | Abe, S, Ito, N, Maity, B, Lu, C, Lu, D, Ueno, T. | | Deposit date: | 2019-02-05 | | Release date: | 2019-12-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Coordination design of cadmium ions at the 4-fold axis channel of the apo-ferritin cage.
Dalton Trans, 48, 2019
|
|
3BOJ
 
 | | Carbonic anhydrase from marine diatom Thalassiosira weissflogii- cadmium bound domain 1 without bound metal (CDCA1-R1) | | Descriptor: | ACETATE ION, Cadmium-specific carbonic anhydrase | | Authors: | Xu, Y, Feng, L, Jeffrey, P.D, Shi, Y, Morel, F.M.M. | | Deposit date: | 2007-12-17 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and metal exchange in the cadmium carbonic anhydrase of marine diatoms.
Nature, 452, 2008
|
|
8PHE
 
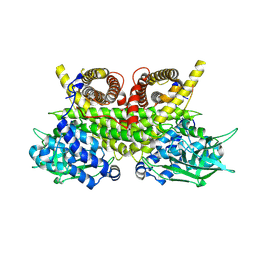 | | ACAD9-WT in complex with ECSIT-CTER | | Descriptor: | Complex I assembly factor ACAD9, mitochondrial, Evolutionarily conserved signaling intermediate in Toll pathway | | Authors: | McGregor, L, Acajjaoui, S, Desfosses, A, Saidi, M, Bacia-Verloop, M, Schwarz, J.J, Juyoux, P, Von Velsen, J, Bowler, M.W, McCarthy, A, Kandiah, E, Gutsche, I, Soler-Lopez, M. | | Deposit date: | 2023-06-19 | | Release date: | 2024-01-24 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The assembly of the Mitochondrial Complex I Assembly complex uncovers a redox pathway coordination.
Nat Commun, 14, 2023
|
|
4XHZ
 
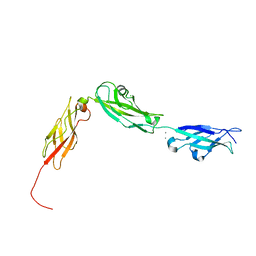 | |
3PPE
 
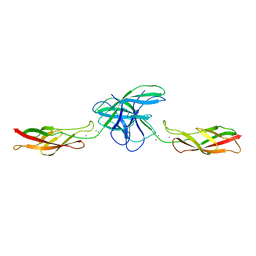 | | Crystal structure of chicken VE-cadherin EC1-2 | | Descriptor: | CALCIUM ION, Vascular endothelial cadherin | | Authors: | Brasch, J, Harrison, O.J, Ahlsen, G, Carnally, S.M, Henderson, R.M, Honig, B, Shapiro, L.S. | | Deposit date: | 2010-11-24 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and binding mechanism of vascular endothelial cadherin: a divergent classical cadherin.
J.Mol.Biol., 408, 2011
|
|
3L6Y
 
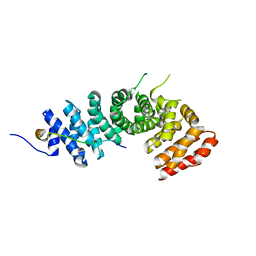 | | Crystal structure of p120 catenin in complex with E-cadherin | | Descriptor: | Catenin delta-1, E-cadherin | | Authors: | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | Deposit date: | 2009-12-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
3L6X
 
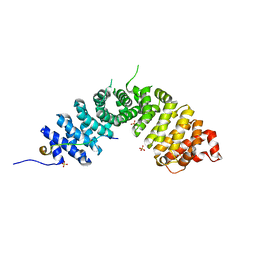 | | Crystal structure of p120 catenin in complex with E-cadherin | | Descriptor: | Catenin delta-1, E-cadherin, SULFATE ION | | Authors: | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | Deposit date: | 2009-12-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
5KJ4
 
 | |
5IU9
 
 | |
6BXZ
 
 | | Crystal Structure of Pig Protocadherin-15 EC10-MAD12 | | Descriptor: | CALCIUM ION, Protocadherin related 15 | | Authors: | De-la-Torre, P, Araya-Secchi, R, Choudhary, D, Sotomayor, M. | | Deposit date: | 2017-12-19 | | Release date: | 2018-11-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A Mechanically Weak Extracellular Membrane-Adjacent Domain Induces Dimerization of Protocadherin-15.
Biophys. J., 115, 2018
|
|
