6GCJ
 
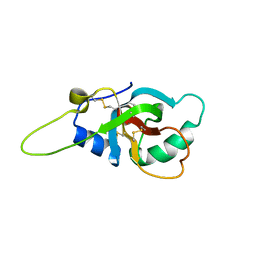 | | Solution structure of the RodA hydrophobin from Aspergillus fumigatus | | Descriptor: | Hydrophobin | | Authors: | Pille, A, Kwan, A, Aimanianda, V, Latge, J.-P, Sunde, M, Guijarro, J.I. | | Deposit date: | 2018-04-18 | | Release date: | 2019-03-27 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Assembly and disassembly of Aspergillus fumigatus conidial rodlets
Cell Surf, 2019
|
|
8Q71
 
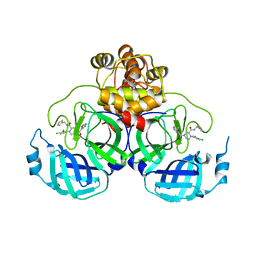 | | Crystal structure of SARS-CoV-2 main protease (MPro) in complex with the inhibitor GC-67 | | Descriptor: | (2~{S})-1-(3,4-dichlorophenyl)-4-(4-methoxypyridin-3-yl)carbonyl-~{N}-(thiophen-2-ylmethyl)piperazine-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Strater, N, Muller, C.E, Sylvester, K, Weisse, R.H, Useini, A, Gao, S, Song, L, Liu, Z, Zhan, P. | | Deposit date: | 2023-08-15 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.322 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Trisubstituted Piperazine Derivatives as Noncovalent Severe Acute Respiratory Syndrome Coronavirus 2 Main Protease Inhibitors with Improved Antiviral Activity and Favorable Druggability.
J.Med.Chem., 66, 2023
|
|
8VF6
 
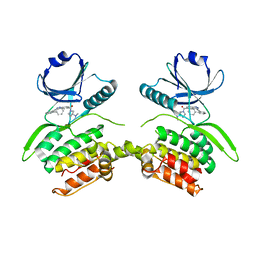 | | Crystal structure of Serine/threonine-protein kinase 33 (STK33) Kinase Domain in complex with inhibitor CDD-2211 | | Descriptor: | Serine/threonine-protein kinase 33, {3-[([1,1'-biphenyl]-2-yl)ethynyl]-1H-indazol-5-yl}[(3R)-3-(dimethylamino)pyrrolidin-1-yl]methanone | | Authors: | Ta, H.M, Kim, C, Ku, K.A, Matzuk, M.M. | | Deposit date: | 2023-12-21 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Reversible male contraception by targeted inhibition of serine/threonine kinase 33.
Science, 384, 2024
|
|
8PIR
 
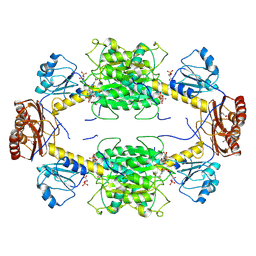 | | Crystal structure of Ser33 in complex with 3-PGA (3-phosphoglycerate) | | Descriptor: | 1,4-BUTANEDIOL, 3-PHOSPHOGLYCERIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Perrone, S, Cifuente, J.O, Marina, A, Mastrella, L, Trastoy, B, Linster, C.L, Guerin, M.E. | | Deposit date: | 2023-06-22 | | Release date: | 2025-01-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Ser33 in complex with 3-PGA (3-phosphoglycerate)
To Be Published
|
|
9QZE
 
 | |
3GLP
 
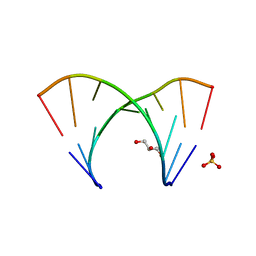 | | 1.23 A resolution X-ray structure of (GCUGCUGC)2 | | Descriptor: | 5'-R(*GP*CP*UP*GP*CP*UP*GP*C)-3', GLYCEROL, SULFATE ION | | Authors: | Kiliszek, A, Kierzek, R, Krzyzosiak, W.J, Rypniewski, W. | | Deposit date: | 2009-03-12 | | Release date: | 2009-05-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structural insights into CUG repeats containing the 'stretched U-U wobble': implications for myotonic dystrophy.
Nucleic Acids Res., 37, 2009
|
|
9IL3
 
 | |
9C6T
 
 | |
6S9P
 
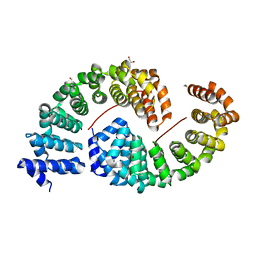 | | Designed Armadillo Repeat protein internal Lock2 fused to target peptide KRKAKITWKR | | Descriptor: | 1,2-ETHANEDIOL, internal Lock2 fused to target peptide KRKAKITWKR | | Authors: | Ernst, P, Zosel, F, Reichen, C, Schuler, B, Pluckthun, A. | | Deposit date: | 2019-07-15 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-Guided Design of a Peptide Lock for Modular Peptide Binders.
Acs Chem.Biol., 15, 2020
|
|
8J6V
 
 | |
9BU4
 
 | | Crystal structure of an MKP5 mutant, Y435W, in complex with an allosteric inhibitor | | Descriptor: | 3,3-dimethyl-1-{[9-(methylsulfanyl)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl]sulfanyl}butan-2-one, Dual specificity protein phosphatase 10 | | Authors: | Manjula, R, Bennett, A.M, Lolis, E. | | Deposit date: | 2024-05-16 | | Release date: | 2025-05-28 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dynamic and structural insights into allosteric regulation on MKP5 a dual-specificity phosphatase.
Nat Commun, 16, 2025
|
|
8KIH
 
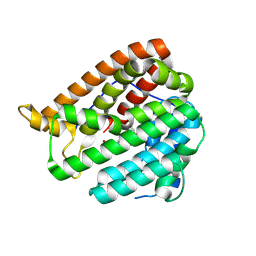 | | PhmA, a type I diterpene synthase without NST/DTE motif | | Descriptor: | (2Z,6E,10E)-2-fluoro-3,7,11,15-tetramethylhexadeca-2,6,10,14-tetraen-1-yl trihydrogen diphosphate, MAGNESIUM ION, diterpene synthase, ... | | Authors: | Zhang, B, Ge, H.M, Zhu, A, Zhang, Y. | | Deposit date: | 2023-08-23 | | Release date: | 2023-10-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biosynthesis of Phomactin Platelet Activating Factor Antagonist Requires a Two-Enzyme Cascade.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7BWQ
 
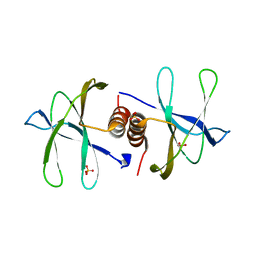 | | Structure of nonstructural protein Nsp9 from SARS-CoV-2 | | Descriptor: | Nsp9, SULFATE ION | | Authors: | Zhang, C, Chen, Y, Li, L, Su, D. | | Deposit date: | 2020-04-15 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.954 Å) | | Cite: | Structural basis for the multimerization of nonstructural protein nsp9 from SARS-CoV-2.
Mol Biomed, 1, 2020
|
|
8T36
 
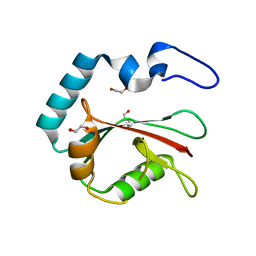 | | Crystal structure of K49 acetylated LC3A in complex with the LIR of TP53INP2/DOR | | Descriptor: | 1,2-ETHANEDIOL, Tumor protein p53-inducible nuclear protein 2,Microtubule-associated proteins 1A/1B light chain 3A chimera | | Authors: | Ali, M.G.H, Wahba, H.M, Cyr, N, Omichinski, J.G. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Structural and functional characterization of the role of acetylation on the interactions of the human Atg8-family proteins with the autophagy receptor TP53INP2/DOR.
Autophagy, 20, 2024
|
|
8OK9
 
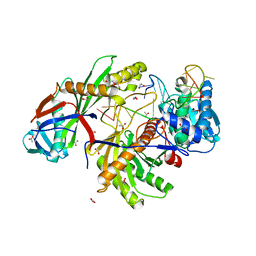 | |
6G82
 
 | |
6FMR
 
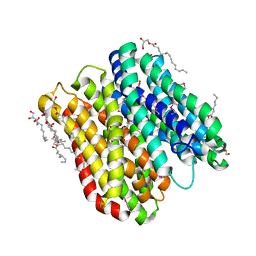 | | IMISX-EP of Se-PepTSt | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Di-or tripeptide:H+ symporter, PHOSPHATE ION | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
7F44
 
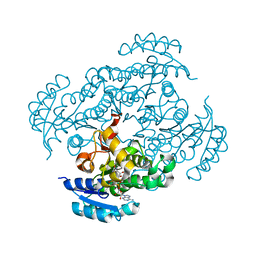 | | Crystal structure of Moraxella catarrhalis enoyl-ACP-reductase (FabI) in complex with the cofactor NAD | | Descriptor: | CALCIUM ION, Enoyl-[acyl-carrier-protein] reductase [NADH], GLYCEROL, ... | | Authors: | Katiki, M, Neetu, N, Pratap, S, Kumar, P. | | Deposit date: | 2021-06-17 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Biochemical and structural basis for Moraxella catarrhalis enoyl-acyl carrier protein reductase (FabI) inhibition by triclosan and estradiol.
Biochimie, 198, 2022
|
|
6M87
 
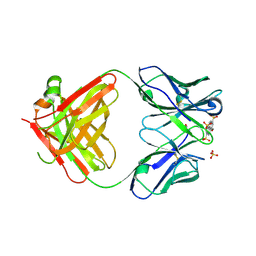 | | Fab 10A6 in complex with MPTS | | Descriptor: | 8-methoxypyrene-1,3,6-trisulfonic acid, Fab 10A6 heavy chain, Fab 10A6 light chain, ... | | Authors: | Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2018-08-21 | | Release date: | 2019-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.609 Å) | | Cite: | Structure and Dynamics of Stacking Interactions in an Antibody Binding Site.
Biochemistry, 58, 2019
|
|
6IYF
 
 | | Structure of pSTING complex | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-12-15 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.764 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
9FMQ
 
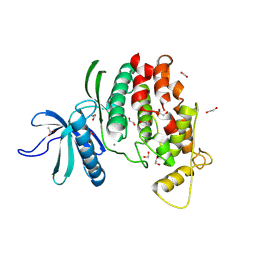 | | Crystal structure of C. merolae LAMMER-like dual specificity kinase (CmLIK) hairpin mutant kinase domain | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Dimos-Roehl, B, Haltenhof, T, Kotte, A, Heyd, F, Loll, B. | | Deposit date: | 2024-06-06 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of C. merolae LAMMER-like dual specificity kinase CmLIK kinase domain
to be published
|
|
1NP9
 
 | |
6YMY
 
 | | Cytochrome c oxidase from Saccharomyces cerevisiae | | Descriptor: | (2R,5S,11R,14R)-5,8,11-trihydroxy-2-(nonanoyloxy)-5,11-dioxido-16-oxo-14-[(propanoyloxy)methyl]-4,6,10,12,15-pentaoxa-5,11-diphosphanonadec-1-yl undecanoate, 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, COPPER (II) ION, ... | | Authors: | Berndtsson, J, Rathore, S, Ott, M. | | Deposit date: | 2020-04-10 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Respiratory supercomplexes enhance electron transport by decreasing cytochrome c diffusion distance.
Embo Rep., 21, 2020
|
|
6VXT
 
 | | Activated Nitrogenase MoFe-protein from Azotobacter vinelandii | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (III) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Kang, W, Hu, Y, Ribbe, M.W. | | Deposit date: | 2020-02-24 | | Release date: | 2020-06-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural evidence for a dynamic metallocofactor during N2reduction by Mo-nitrogenase.
Science, 368, 2020
|
|
5J9M
 
 | |
