1DMX
 
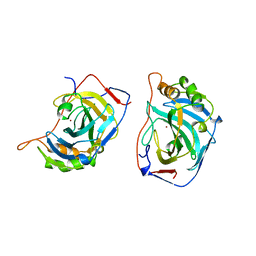 | |
1YAM
 
 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
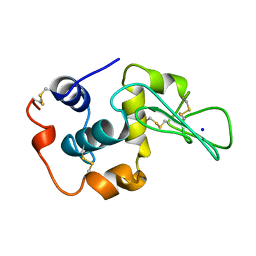
1YAN
 
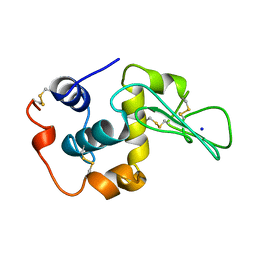 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
1YAQ
 
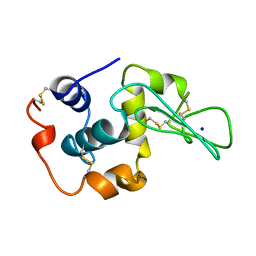 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
1YAP
 
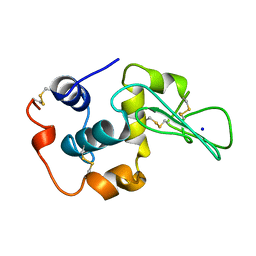 | | CONTRIBUTION OF HYDROPHOBIC RESIDUES TO THE STABILITY OF HUMAN LYSOZYME: CALORIMETRIC STUDIES AND X-RAY STRUCTURAL ANALYSIS OF THE FIVE ISOLEUCINE TO VALINE MUTANTS | | Descriptor: | LYSOZYME, SODIUM ION | | Authors: | Yamagata, Y, Kaneda, H, Fujii, S, Takano, K, Ogasahara, K, Kanaya, E, Kikuchi, M, Oobatake, M, Yutani, K. | | Deposit date: | 1995-09-29 | | Release date: | 1996-04-03 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Contribution of hydrophobic residues to the stability of human lysozyme: calorimetric studies and X-ray structural analysis of the five isoleucine to valine mutants.
J.Mol.Biol., 254, 1995
|
|
1HSI
 
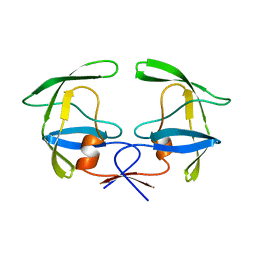 | |
1HSG
 
 | |
1FRS
 
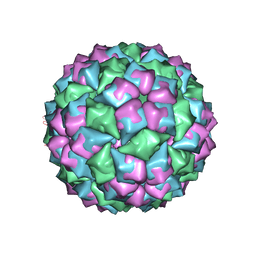 | |
1HSH
 
 | |
1XAC
 
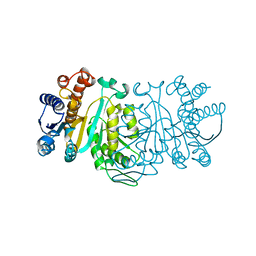 | |
1XAD
 
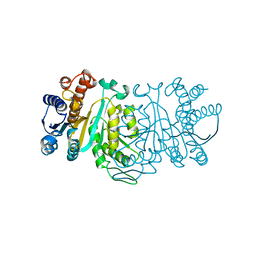 | |
1XAB
 
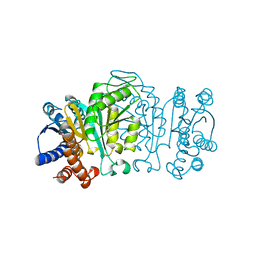 | |
1XAA
 
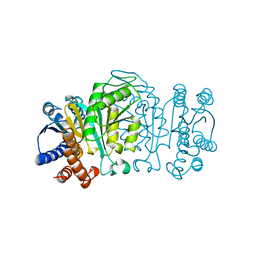 | |
1YMU
 
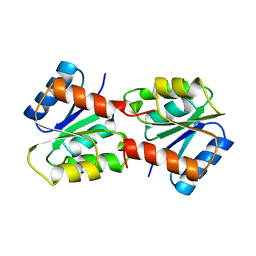 | |
1YMV
 
 | |
1LEY
 
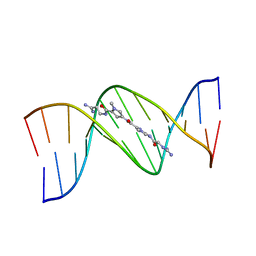 | | STRUCTURE OF A DICATIONIC MONOIMIDAZOLE LEXITROPSIN BOUND TO DNA (ORIENTATION 2) | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3'), MONOIMIDAZOLE LEXITROPSIN | | Authors: | Goodsell, D.S, Ng, H.L, Kopka, M.L, Lown, J.W, Dickerson, R.E. | | Deposit date: | 1995-10-10 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of a dicationic monoimidazole lexitropsin bound to DNA.
Biochemistry, 34, 1995
|
|
1IRO
 
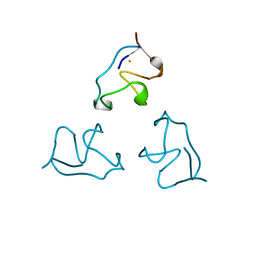 | | RUBREDOXIN (OXIDIZED, FE(III)) AT 1.1 ANGSTROMS RESOLUTION | | Descriptor: | FE (III) ION, RUBREDOXIN | | Authors: | Dauter, Z, Wilson, K.S, Sieker, L.C, Moulis, J.M, Meyer, J. | | Deposit date: | 1995-12-13 | | Release date: | 1996-04-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Zinc- and iron-rubredoxins from Clostridium pasteurianum at atomic resolution: a high-precision model of a ZnS4 coordination unit in a protein.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
2PLH
 
 | | STRUCTURE OF ALPHA-1-PUROTHIONIN AT ROOM TEMPERATURE AND 2.8 ANGSTROMS RESOLUTION | | Descriptor: | 2-BUTANOL, ACETATE ION, ALPHA-1-PUROTHIONIN, ... | | Authors: | Teeter, M.M, Stec, B, Rao, U. | | Deposit date: | 1993-07-09 | | Release date: | 1996-04-03 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Refinement of purothionins reveals solute particles important for lattice formation and toxicity. Part 1: alpha1-purothionin revisited.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1MUT
 
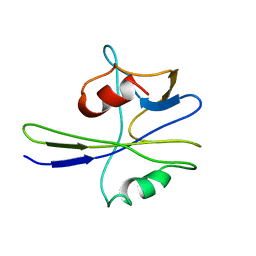 | | NMR STUDY OF MUTT ENZYME, A NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE | | Descriptor: | NUCLEOSIDE TRIPHOSPHATE PYROPHOSPHOHYDROLASE | | Authors: | Abeygunawardana, C, Weber, D.J, Gittis, A.G, Frick, D.N, Lin, J, Miller, A.-F, Bessman, M.J, Mildvan, A.S. | | Deposit date: | 1995-09-14 | | Release date: | 1996-04-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the MutT enzyme, a nucleoside triphosphate pyrophosphohydrolase.
Biochemistry, 34, 1995
|
|
1GFF
 
 | |
1OSU
 
 | |
252D
 
 | |
1RLB
 
 | |
1PTH
 
 | | The Structural Basis of Aspirin Activity Inferred from the Crystal Structure of Inactivated Prostaglandin H2 Synthase | | Descriptor: | 2-HYDROXYBENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Loll, P.J, Picot, D, Garavito, R.M. | | Deposit date: | 1995-04-11 | | Release date: | 1996-04-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The structural basis of aspirin activity inferred from the crystal structure of inactivated prostaglandin H2 synthase.
Nat.Struct.Biol., 2, 1995
|
|
1ZQE
 
 | |
