2W02
 
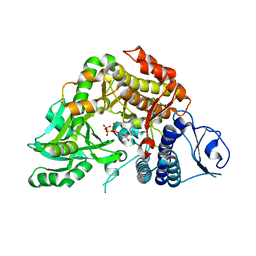 | | Co-complex Structure of Achromobactin Synthetase Protein D (AcsD) with ATP from Pectobacterium Chrysanthemi | | Descriptor: | ACSD, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Schmelz, S, McMahon, S.A, Kadi, N, Song, L, Oves-Costales, D, Oke, M, Liu, H, Johnson, K.A, Carter, L, White, M.F, Challis, G.L, Naismith, J.H. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | AcsD catalyzes enantioselective citrate desymmetrization in siderophore biosynthesis.
Nat. Chem. Biol., 5, 2009
|
|
4IPC
 
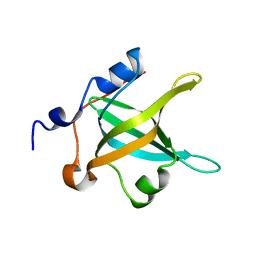 | | Structure of the N-terminal domain of RPA70, E7R mutant | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPD
 
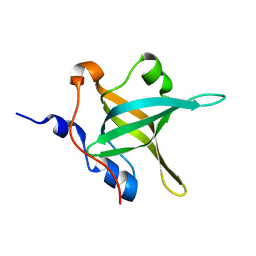 | | Structure of the N-terminal domain of RPA70, E100R mutant | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPG
 
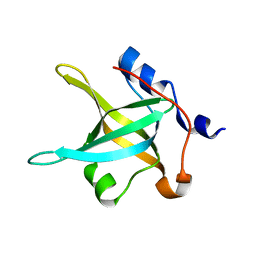 | | Structure of the N-terminal domain of RPA70, E7R, E100R mutant | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
4IPH
 
 | | Structure of N-terminal domain of RPA70 in complex with VU079104 inhibitor | | Descriptor: | Replication protein A 70 kDa DNA-binding subunit, ~{N}-(2,3-dimethylphenyl)-7-oxidanylidene-12-sulfanylidene-5,11-dithia-1,8-diazatricyclo[7.3.0.0^{2,6}]dodeca-2(6),3,9-triene-10-carboxamide | | Authors: | Feldkamp, M.D, Frank, A.O, Vangamudi, B, Fesik, S.W, Chazin, W.J. | | Deposit date: | 2013-01-09 | | Release date: | 2013-09-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Surface Reengineering of RPA70N Enables Cocrystallization with an Inhibitor of the Replication Protein A Interaction Motif of ATR Interacting Protein.
Biochemistry, 52, 2013
|
|
3OUQ
 
 | | Structure of N-terminal hexaheme fragment of GSU1996 | | Descriptor: | Cytochrome c family protein, HEME C | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2010-09-15 | | Release date: | 2010-12-29 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of a novel dodecaheme cytochrome c from Geobacter sulfurreducens reveals an extended 12nm protein with interacting hemes.
J.Struct.Biol., 174, 2011
|
|
3OV0
 
 | | Structure of dodecaheme cytochrome c GSU1996 | | Descriptor: | Cytochrome c family protein, HEME C | | Authors: | Pokkuluri, P.R, Schiffer, M. | | Deposit date: | 2010-09-15 | | Release date: | 2010-12-29 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of a novel dodecaheme cytochrome c from Geobacter sulfurreducens reveals an extended 12nm protein with interacting hemes.
J.Struct.Biol., 174, 2011
|
|
2KT4
 
 | | Lipocalin Q83 is a Siderocalin | | Descriptor: | Extracellular fatty acid-binding protein, GALLIUM (III) ION, N,N',N''-[(3S,7S,11S)-2,6,10-trioxo-1,5,9-trioxacyclododecane-3,7,11-triyl]tris(2,3-dihydroxybenzamide) | | Authors: | Coudevylle, N, Geist, L, Hartl, M, Kontaxis, G, Bister, K, Konrat, R. | | Deposit date: | 2010-01-18 | | Release date: | 2010-09-08 | | Last modified: | 2016-01-27 | | Method: | SOLUTION NMR | | Cite: | The v-myc-induced Q83 lipocalin is a siderocalin.
J.Biol.Chem., 285, 2010
|
|
2LBV
 
 | | Siderocalin Q83 reveals a dual ligand binding mode | | Descriptor: | ARACHIDONIC ACID, Extracellular fatty acid-binding protein, GALLIUM (III) ION, ... | | Authors: | Coudevylle, N, Hoetzinger, M, Geist, L, Kontaxis, G, Bister, K, Konrat, R. | | Deposit date: | 2011-04-07 | | Release date: | 2012-02-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Lipocalin Q83 reveals a dual ligand binding mode with potential implications for the functions of siderocalins
Biochemistry, 50, 2011
|
|
1CLQ
 
 | |
3CMP
 
 | |
7Q4D
 
 | | Local refinement structure of the two interacting N-domains of full-length, dimeric, soluble somatic angiotensin I-converting enzyme | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lubbe, L, Sewell, B.T, Sturrock, E.D. | | Deposit date: | 2021-10-30 | | Release date: | 2022-07-20 | | Last modified: | 2022-08-24 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Cryo-EM reveals mechanisms of angiotensin I-converting enzyme allostery and dimerization.
Embo J., 41, 2022
|
|
3ERD
 
 | | HUMAN ESTROGEN RECEPTOR ALPHA LIGAND-BINDING DOMAIN IN COMPLEX WITH DIETHYLSTILBESTROL AND A GLUCOCORTICOID RECEPTOR INTERACTING PROTEIN 1 NR BOX II PEPTIDE | | Descriptor: | ACETIC ACID, CHLORIDE ION, DIETHYLSTILBESTROL, ... | | Authors: | Shiau, A.K, Barstad, D, Loria, P.M, Cheng, L, Kushner, P.J, Agard, D.A, Greene, G.L. | | Deposit date: | 1999-03-31 | | Release date: | 1999-04-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen.
Cell(Cambridge,Mass.), 95, 1998
|
|
2IRM
 
 | | Crystal structure of mitogen-activated protein kinase kinase kinase 7 interacting protein 1 from Anopheles gambiae | | Descriptor: | mitogen-activated protein kinase kinase kinase 7 interacting protein 1 | | Authors: | Jin, X, Bonanno, J.B, Pelletier, L, Freeman, J.C, Wasserman, S, Sauder, J.M, Burley, S.K, Shapiro, L, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-10-15 | | Release date: | 2006-11-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural genomics of protein phosphatases.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
2QAC
 
 | | The closed MTIP-MyosinA-tail complex from the malaria parasite invasion machinery | | Descriptor: | Myosin A tail domain interacting protein MTIP, Myosin-A | | Authors: | Bosch, J, Turley, S, Roach, C.M, Daly, T.M, Bergman, L.W, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2007-06-14 | | Release date: | 2007-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Closed MTIP-Myosin A-Tail Complex from the Malaria Parasite Invasion Machinery.
J.Mol.Biol., 372, 2007
|
|
5HP2
 
 | | Human Adenosine Deaminase Acting on dsRNA (ADAR2) bound to dsRNA sequence derived from S. cerevisiae BDF2 gene with AU basepair at reaction site | | Descriptor: | Double-stranded RNA-specific editase 1, INOSITOL HEXAKISPHOSPHATE, RNA (5'-R(*GP*AP*CP*UP*GP*AP*AP*CP*GP*AP*CP*UP*AP*AP*UP*GP*UP*GP*GP*GP*GP*AP*A)-3'), ... | | Authors: | Matthews, M.M, Fisher, A.J, Beal, P.A. | | Deposit date: | 2016-01-20 | | Release date: | 2016-04-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.983 Å) | | Cite: | Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity.
Nat.Struct.Mol.Biol., 23, 2016
|
|
4L0W
 
 | |
3IY0
 
 | | Variable domains of the x-ray structure of Fab 14 fitted into the cryoEM reconstruction of the virus-Fab 14 complex | | Descriptor: | Fab 14, heavy domain, light domain | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-07 | | Release date: | 2009-05-12 | | Last modified: | 2011-07-13 | | Method: | ELECTRON MICROSCOPY (12.5 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IY7
 
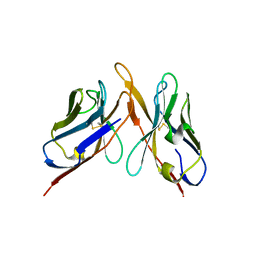 | | Variable domains of the computer generated model (WAM) of Fab F fitted into the cryoEM reconstruction of the virus-Fab F complex | | Descriptor: | fragment from neutralizing antibody F (heavy chain), fragment from neutralizing antibody F (light chain) | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (14 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IY2
 
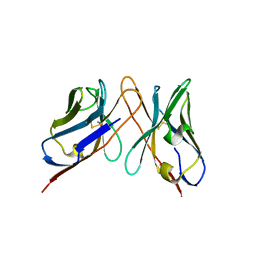 | | Variable domains of the computer generated model (WAM) of Fab 6 fitted into the cryoEM reconstruction of the virus-Fab 6 complex | | Descriptor: | Antibody 6, heavy chain, light chain | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IY4
 
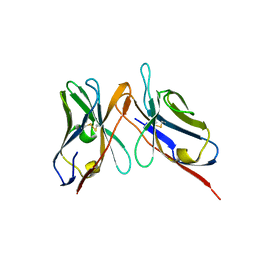 | | Variable domains of the computer generated model (WAM) of Fab 15 fitted into the cryoEM reconstruction of the virus-Fab 15 complex | | Descriptor: | fragment of neutralizing antibody 15 (heavy chain), fragment of neutralizing antibody 15 (light chain) | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (11.7 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IY6
 
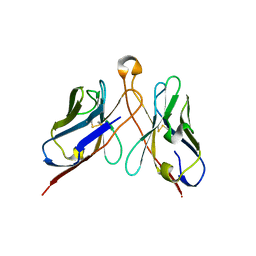 | | Variable domains of the computer generated model (WAM) of Fab E fitted into the cryoEM reconstruction of the virus-Fab E complex | | Descriptor: | fragment from neutralizing antibody E (heavy chain), fragment from neutralizing antibody E (light chain) | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IY1
 
 | | Variable domains of the WAM of Fab B fitted into the cryoEM reconstruction of the virus-Fab B complex | | Descriptor: | Fab B, heavy chain, light chain | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IY5
 
 | | Variable domains of the mouse Fab (1AIF) fitted into the cryoEM reconstruction of the virus-Fab 16 complex | | Descriptor: | antibody fragment IGG2A (heavy chain), antibody fragment IGG2A (light chain) | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (18 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
3IY3
 
 | | Variable domains of the computer generated model (WAM) of Fab 8 fitted into the cryoEM reconstruction of the virus-Fab 8 complex | | Descriptor: | antibody fragment from neutralizing antibody 8 (heavy chain), antibody fragment from neutralizing antibody 8 (light chain) | | Authors: | Hafenstein, S, Bowman, V.D, Sun, T, Nelson, C.D, Palermo, L.M, Chipman, P.R, Battisti, A.J, Parrish, C.R, Rossmann, M.G. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (11.1 Å) | | Cite: | Structural comparison of different antibodies interacting with parvovirus capsids
J.Virol., 83, 2009
|
|
