1PQI
 
 | | T4 LYSOZYME CORE REPACKING MUTANT I118L/CORE7/TA | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme, ... | | Authors: | Mooers, B.H, Datta, D, Baase, W.A, Zollars, E.S, Mayo, S.L, Matthews, B.W. | | Deposit date: | 2003-06-18 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Repacking the Core of T4 Lysozyme by Automated Design
J.Mol.Biol., 332, 2003
|
|
3ZDD
 
 | | Structure of E. coli ExoIX in complex with the palindromic 5ov6 oligonucleotide and potassium | | Descriptor: | 1,2-ETHANEDIOL, 5OV6 DNA, ISOPROPYL ALCOHOL, ... | | Authors: | Flemming, C.S, Hemsworth, G.R, Anstey-Gilbert, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The structure of Escherichia coli ExoIX--implications for DNA binding and catalysis in flap endonucleases.
Nucleic Acids Res., 41, 2013
|
|
1PQU
 
 | | Crystal Structure of the H277N Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae Bound with NADP, S-methyl cysteine sulfoxide and cacodylate | | Descriptor: | Aspartate-semialdehyde dehydrogenase, CACODYLATE ION, CYSTEINE, ... | | Authors: | Blanco, J, Moore, R.A, Viola, R.E. | | Deposit date: | 2003-06-19 | | Release date: | 2004-08-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2ZC1
 
 | | Organophosphorus Hydrolase from Deinococcus radiodurans | | Descriptor: | BROMIDE ION, COBALT (II) ION, Phosphotriesterase | | Authors: | Larsen, S.D, Hawwa, R, Ratia, K, Santarsiero, B.D, Mesecar, A.D. | | Deposit date: | 2007-11-02 | | Release date: | 2008-11-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-Ray Structural Insights into a Phosphotriesterase
to be published
|
|
4C5Z
 
 | | Crystal structure of A. niger ochratoxinase | | Descriptor: | OCHRATOXINASE | | Authors: | Dobritzsch, D, Wang, H, Schneider, G, Yu, S. | | Deposit date: | 2013-09-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Characterization of Ochratoxinase, a Novel Mycotoxin Degrading Enzyme.
Biochem.J., 462, 2014
|
|
1NXQ
 
 | | Crystal Structure of R-alcohol dehydrogenase (RADH) (apoenyzme) from Lactobacillus brevis | | Descriptor: | MAGNESIUM ION, R-alcohol dehydrogenase | | Authors: | Niefind, K, Muller, J, Riebel, B, Hummel, W, Schomburg, D. | | Deposit date: | 2003-02-11 | | Release date: | 2003-04-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The crystal structure of R-specific alcohol dehydrogenase from Lactobacillus brevis suggests the structural basis of its metal dependency
J.Mol.Biol., 327, 2003
|
|
2ED4
 
 | | Crystal structure of flavin reductase HpaC complexed with FAD and NAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, flavin reductase (HpaC) of 4-hydroxyphenylacetate 3-monooxygenae | | Authors: | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-02-14 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
3ZD8
 
 | | Potassium bound structure of E. coli ExoIX in P1 | | Descriptor: | POTASSIUM ION, PROTEIN XNI | | Authors: | Anstey-Gilbert, C.S, Hemsworth, G.R, Flemming, C.S, Hodskinson, M.R.G, Zhang, J, Sedelnikova, S.E, Stillman, T.J, Sayers, J.R, Artymiuk, P.J. | | Deposit date: | 2012-11-26 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of E. Coli Exoix - Implications for DNA Binding and Catalysis in Flap Endonucleases
Nucleic Acids Res., 41, 2013
|
|
1YWS
 
 | |
3ZF0
 
 | | Phage dUTPases control transfer of virulence genes by a proto-oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase D81A mutant with dUpNHpp). | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPASE, NICKEL (II) ION | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
4C97
 
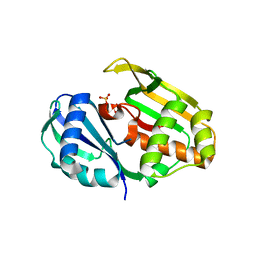 | | Cas6 (TTHA0078) H37A mutant | | Descriptor: | CAS6A, SULFATE ION | | Authors: | Jinek, M, Niewoehner, O, Doudna, J.A. | | Deposit date: | 2013-10-02 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of Crispr RNA Recognition and Processing by Cas6 Endonucleases.
Nucleic Acids Res., 42, 2014
|
|
1O01
 
 | | Human mitochondrial aldehyde dehydrogenase complexed with crotonaldehyde, NAD(H) and Mg2+ | | Descriptor: | (2E)-BUT-2-ENAL, 1,2-ETHANEDIOL, Aldehyde dehydrogenase, ... | | Authors: | Perez-Miller, S.J, Hurley, T.D. | | Deposit date: | 2003-02-20 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Coenzyme isomerization is integral to catalysis in aldehyde dehydrogenase
Biochemistry, 42, 2003
|
|
4CAZ
 
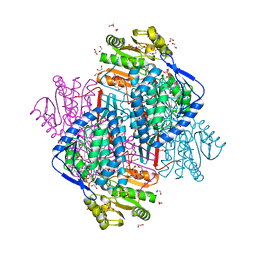 | | CRYSTAL STRUCTURE OF BETAINE ALDEHYDE DEHYDROGENASE FROM Pseudomonas aeruginosa IN COMPLEX WITH NADH | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, BETAINE ALDEHYDE DEHYDROGENASE, ... | | Authors: | Gonzalez-Segura, L, Diaz-Sanchez, A.G, Rodriguez-Sotres, R, Mujica-Jimenez, C, Munoz-Clares, R.A. | | Deposit date: | 2013-10-09 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Structural Bases of the Dual Coenzyme Specificity of Betaine Aldehyde Dehydrogenase from Pseudomonas Aeruginosa
To be Published
|
|
1NZZ
 
 | |
4GO3
 
 | |
1O20
 
 | |
1YYT
 
 | | D100E Trichodiene Synthase: Complex With Mg, Pyrophosphate, and (4R)-7-azabisabolene | | Descriptor: | (1S)-N,4-DIMETHYL-N-(4-METHYLPENT-3-ENYL)CYCLOHEX-3-ENAMINIUM, MAGNESIUM ION, PYROPHOSPHATE 2-, ... | | Authors: | Vedula, L.S, Rynkiewicz, M.J, Pyun, H.J, Coates, R.M, Cane, D.E, Christianson, D.W. | | Deposit date: | 2005-02-25 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Molecular Recognition of the Substrate Diphosphate Group Governs Product Diversity in Trichodiene Synthase Mutants.
Biochemistry, 44, 2005
|
|
2EHQ
 
 | | Crystal analysis of 1-pyrroline-5-carboxylate dehydrogenase from thermus with bound NADP | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1-pyrroline-5-carboxylate dehydrogenase, ACETATE ION, ... | | Authors: | Inagaki, E, Sakamoto, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-07 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | New insights into the binding mode of coenzymes: structure of Thermus thermophilus Delta1-pyrroline-5-carboxylate dehydrogenase complexed with NADP+.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2ZCV
 
 | | Crystal structure of NADPH-dependent quinone oxidoreductase QOR2 complexed with NADPH from escherichia coli | | Descriptor: | COPPER (II) ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Kim, I.K, Yim, H.S, Kim, M.K, Kim, D.W, Kim, Y.M, Cha, S.S, Kang, S.O. | | Deposit date: | 2007-11-13 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a new type of NADPH-dependent quinone oxidoreductase (QOR2) from Escherichia coli
J.Mol.Biol., 379, 2008
|
|
4BO7
 
 | | Crystal structure of 3-oxoacyl-(acyl-carrier-protein) reductase (FabG) from Pseudomonas aeruginosa in complex with N-(2,3-dihydro-1H-inden- 5-yl)tetrazolo(1,5-b)pyridazin-6-amine at 2.6A resolution | | Descriptor: | 3-OXOACYL-[ACYL-CARRIER-PROTEIN] REDUCTASE FABG, N-(2,3-dihydro-1H-inden-5-yl)tetrazolo[1,5-b]pyridazin-6-amine | | Authors: | Cukier, C.D, Schnell, R, Lindqvist, Y, Schneider, G. | | Deposit date: | 2013-05-18 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of an Allosteric Inhibitor Binding Site in 3-Oxo-Acyl-Acp Reductase from Pseudomonas Aeruginosa
Acs Chem.Biol., 8, 2013
|
|
2ZEY
 
 | | Family 16 carbohydrate binding module | | Descriptor: | CALCIUM ION, S-layer associated multidomain endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose, ... | | Authors: | Nair, S.K, Bae, B. | | Deposit date: | 2007-12-18 | | Release date: | 2008-03-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular Basis for the Selectivity and Specificity of Ligand Recognition by the Family 16 Carbohydrate-binding Modules from Thermoanaerobacterium polysaccharolyticum ManA
J.Biol.Chem., 283, 2008
|
|
4GSM
 
 | |
4BTE
 
 | | DJ-1 Cu(I) complex | | Descriptor: | COPPER (I) ION, PROTEIN DJ-1 | | Authors: | Puno, M.R.A, Odell, M, Moody, P.C.E. | | Deposit date: | 2013-06-14 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structure of Cu(I)-Bound Dj-1 Reveals a Biscysteinate Metal Binding Site at the Homodimer Interface: Insights Into Mutational Inactivation of Dj-1 in Parkinsonism.
J.Am.Chem.Soc., 135, 2013
|
|
2EKL
 
 | |
4BVS
 
 | | Cyanuric acid hydrolase: evolutionary innovation by structural concatenation. | | Descriptor: | 1,3,5-triazine-2,4,6-triamine, CYANURIC ACID AMIDOHYDROLASE, MAGNESIUM ION | | Authors: | Peat, T.S, Balotra, S, Wilding, M, French, N.G, Briggs, L.J, Panjikar, S, Cowieson, N, Newman, J, Scott, C. | | Deposit date: | 2013-06-28 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cyanuric Acid Hydrolase: Evolutionary Innovation by Structural Concatenation.
Mol.Microbiol., 88, 2013
|
|
