3ETQ
 
 | |
3E97
 
 | |
3E6D
 
 | | Crystal Structure of CprK C200S | | Descriptor: | Cyclic nucleotide-binding protein | | Authors: | Levy, C. | | Deposit date: | 2008-08-15 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular basis of halorespiration control by CprK, a CRP-FNR type transcriptional regulator
Mol.Microbiol., 70, 2008
|
|
3E6C
 
 | | CprK OCPA DNA Complex | | Descriptor: | (3-CHLORO-4-HYDROXYPHENYL)ACETIC ACID, Cyclic nucleotide-binding protein, DNA (5'-D(P*DGP*DCP*DAP*DTP*DTP*DAP*DAP*DCP*DAP*DTP*DGP*DCP*DC)-3'), ... | | Authors: | Levy, C. | | Deposit date: | 2008-08-15 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular basis of halorespiration control by CprK, a CRP-FNR type transcriptional regulator
Mol.Microbiol., 70, 2008
|
|
3E6B
 
 | | OCPA complexed CprK (C200S) | | Descriptor: | (3-CHLORO-4-HYDROXYPHENYL)ACETIC ACID, Cyclic nucleotide-binding protein | | Authors: | Levy, C. | | Deposit date: | 2008-08-15 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Molecular basis of halorespiration control by CprK, a CRP-FNR type transcriptional regulator
Mol.Microbiol., 70, 2008
|
|
3E5X
 
 | | OCPA complexed CprK | | Descriptor: | (3-CHLORO-4-HYDROXYPHENYL)ACETIC ACID, Cyclic nucleotide-binding protein | | Authors: | Levy, C. | | Deposit date: | 2008-08-14 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Molecular basis of halorespiration control by CprK, a CRP-FNR type transcriptional regulator
Mol.Microbiol., 70, 2008
|
|
3E5U
 
 | | OCPA complexed CprK (C200S) | | Descriptor: | (3-CHLORO-4-HYDROXYPHENYL)ACETIC ACID, Cyclic nucleotide-binding protein, PHOSPHATE ION, ... | | Authors: | Levy, C. | | Deposit date: | 2008-08-14 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Molecular basis of halorespiration control by CprK, a CRP-FNR type transcriptional regulator
Mol.Microbiol., 70, 2008
|
|
3E5Q
 
 | | Unbound Oxidised CprK | | Descriptor: | Cyclic nucleotide-binding protein | | Authors: | Levy, C. | | Deposit date: | 2008-08-14 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Molecular basis of halorespiration control by CprK, a CRP-FNR type transcriptional regulator
Mol.Microbiol., 70, 2008
|
|
3DV8
 
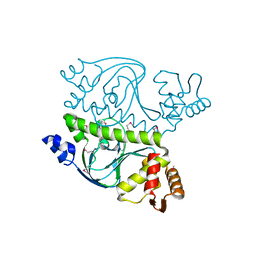 | |
3DKW
 
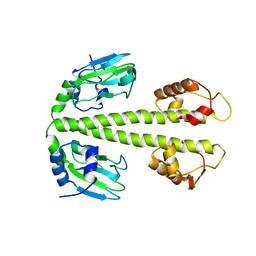 | | Crystal Structure of DNR from Pseudomonas aeruginosa. | | Descriptor: | DNR protein | | Authors: | Giardina, G. | | Deposit date: | 2008-06-26 | | Release date: | 2009-05-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A dramatic conformational rearrangement is necessary for the activation of DNR from Pseudomonas aeruginosa. Crystal structure of wild-type DNR.
Proteins, 2009
|
|
3D0S
 
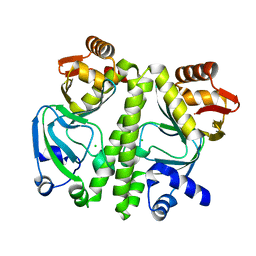 | |
3CO2
 
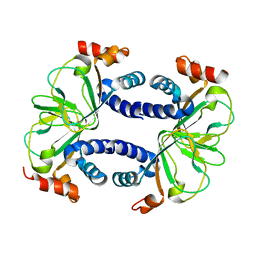 | |
3CLP
 
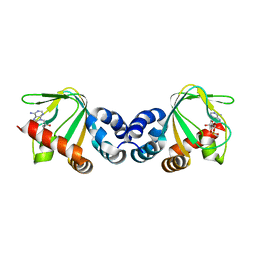 | | M. loti cyclic-nucleotide binding domain mutant 2 | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Mll3241 protein | | Authors: | Clayton, G.M, Altieri, S.L, Thomas, L.R, Morais-Cabral, J.H. | | Deposit date: | 2008-03-19 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Energetic Analysis of Activation by a Cyclic Nucleotide Binding Domain.
J.Mol.Biol., 381, 2008
|
|
3CL1
 
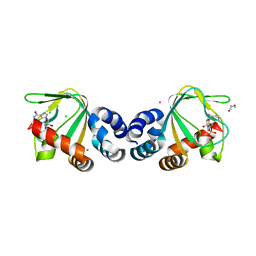 | | M. loti cyclic-nucleotide binding domain, cyclic-GMP bound | | Descriptor: | CHLORIDE ION, CYCLIC GUANOSINE MONOPHOSPHATE, Mll3241 protein, ... | | Authors: | Clayton, G.M, Alteiri, S.L, Thomas, L.R, Morais-Cabral, J.H. | | Deposit date: | 2008-03-18 | | Release date: | 2008-08-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and Energetic Analysis of Activation by a Cyclic Nucleotide Binding Domain.
J.Mol.Biol., 381, 2008
|
|
3BPZ
 
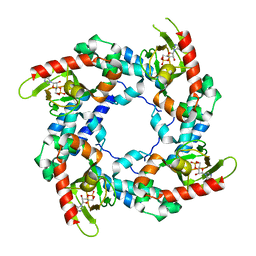 | | HCN2-I 443-460 E502K in the presence of cAMP | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2 | | Authors: | Craven, K.B, Olivier, N.B, Zagotta, W.N. | | Deposit date: | 2007-12-19 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | C-terminal movement during gating in cyclic nucleotide-modulated channels.
J.Biol.Chem., 283, 2008
|
|
3BEH
 
 | |
3B02
 
 | | Crystal structure of TTHB099, a transcriptional regulator CRP family from Thermus thermophilus HB8 | | Descriptor: | Transcriptional regulator, Crp family | | Authors: | Agari, Y, Kuramitsu, S, Shinkai, A, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2011-06-03 | | Release date: | 2011-06-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | X-ray crystal structure of TTHB099, a CRP/FNR superfamily transcriptional regulator from Thermus thermophilus HB8, reveals a DNA-binding protein with no required allosteric effector molecule
Proteins, 80, 2012
|
|
2ZD9
 
 | |
2ZCW
 
 | | Crystal Structure of TTHA1359, a Transcriptional Regulator, CRP/FNR family from Thermus thermophilus HB8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Transcriptional regulator, FNR/CRP family | | Authors: | Agari, Y, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-11-13 | | Release date: | 2007-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Global gene expression mediated by Thermus thermophilus SdrP, a CRP/FNR family transcriptional regulator
Mol.Microbiol., 70, 2008
|
|
2Z69
 
 | |
2XKP
 
 | |
2XKO
 
 | |
2XHK
 
 | |
2XGX
 
 | | Crystal structure of transcription factor NtcA from Synechococcus elongatus (mercury derivative) | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLOBAL NITROGEN REGULATOR, ... | | Authors: | Llacer, J.L, Castells, M.A, Rubio, V. | | Deposit date: | 2010-06-08 | | Release date: | 2010-08-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis for the Regulation of Ntca-Dependent Transcription by Proteins Pipx and Pii.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2WC2
 
 | |
