6DC4
 
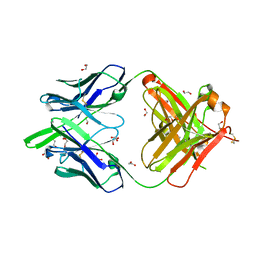 | | RSV-neutralizing human antibody AM22 | | Descriptor: | 1,2-ETHANEDIOL, Fab AM22 Heavy Chain, Fab AM22 Light Chain | | Authors: | Jones, H.G, McLellan, J.S. | | Deposit date: | 2018-05-04 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Alternative conformations of a major antigenic site on RSV F.
Plos Pathog., 15, 2019
|
|
7BI1
 
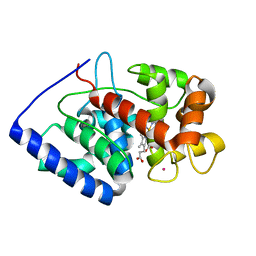 | | XFEL crystal structure of soybean ascorbate peroxidase compound II | | Descriptor: | Ascorbate peroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kwon, H, Tosha, T, Sugimoto, H, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2021-01-12 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | XFEL Crystal Structures of Peroxidase Compound II.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6QV8
 
 | |
7QE5
 
 | |
5IAZ
 
 | |
6QWO
 
 | | Zinc-reconstituted ODP from T. maritima | | Descriptor: | Oxygen-binding diiron protein, ZINC ION | | Authors: | Muok, A.R, Crane, B.R. | | Deposit date: | 2019-03-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A di-iron protein recruited as an Fe[II] and oxygen sensor for bacterial chemotaxis functions by stabilizing an iron-peroxy species.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5M3I
 
 | | Macrodomain of Mycobacterium tuberculosis DarG | | Descriptor: | CHLORIDE ION, RNase III inhibitor | | Authors: | Ariza, A. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | The Toxin-Antitoxin System DarTG Catalyzes Reversible ADP-Ribosylation of DNA.
Mol. Cell, 64, 2016
|
|
6ZZ6
 
 | | Cryo-EM structure of S.cerevisiae cohesin-Scc2-DNA complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (34-MER), MAGNESIUM ION, ... | | Authors: | Lee, B.-G, Gonzalez Llamazares, A, Collier, J, Nasmyth, K.A, Lowe, J. | | Deposit date: | 2020-08-04 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Transport of DNA within cohesin involves clamping on top of engaged heads by Scc2 and entrapment within the ring by Scc3.
Elife, 9, 2020
|
|
6QP2
 
 | | Crystal structure of the PLP-bound C-S lyase from Staphylococcus hominis | | Descriptor: | Aminotransferase, PYRIDOXAL-5'-PHOSPHATE, Polyhistidine tag | | Authors: | Herman, R, Rudden, M, Wilkinson, A.J, Hanai, S, Thomas, G.H. | | Deposit date: | 2019-02-13 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The molecular basis of thioalcohol production in human body odour.
Sci Rep, 10, 2020
|
|
7A76
 
 | |
6ANN
 
 | |
2BF3
 
 | | Crystal structure of a toluene 4-monooxygenase catalytic effector protein variant missing ten N-terminal residues (delta-N10 T4moD) | | Descriptor: | HEPTANE-1,2,3-TRIOL, TOLUENE-4-MONOOXYGENASE SYSTEM PROTEIN D | | Authors: | Lountos, G.T, Mitchell, K.H, Studts, J.M, Fox, B.G, Orville, A.M. | | Deposit date: | 2004-12-03 | | Release date: | 2005-05-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures and Functional Studies of T4Mod, the Toluene 4-Monooxygenase Catalytic Effector Protein
Biochemistry, 44, 2005
|
|
5TH0
 
 | |
5THC
 
 | |
2BF5
 
 | | Crystal structure of a toluene 4-monooxygenase catalytic effector protein variant missing four N-terminal residues (delta-N4 T4moD) | | Descriptor: | TOLUENE-4-MONOOXYGENASE SYSTEM PROTEIN D | | Authors: | Lountos, G.T, Mitchell, K.H, Studts, J.M, Fox, B.G, Orville, A.M. | | Deposit date: | 2004-12-03 | | Release date: | 2005-05-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Crystal Structures and Functional Studies of T4Mod, the Toluene 4-Monooxygenase Catalytic Effector Protein
Biochemistry, 44, 2005
|
|
6DET
 
 | |
6QSS
 
 | | Crystal Structure of Ignicoccus islandicus malate dehydrogenase co-crystallized with 10 mM Tb-Xo4 | | Descriptor: | CHLORIDE ION, Malate dehydrogenase, TERBIUM(III) ION, ... | | Authors: | Roche, J, Girard, E, Madern, D. | | Deposit date: | 2019-02-21 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | SOLUTION SCATTERING (1.892 Å), X-RAY DIFFRACTION | | Cite: | The archaeal LDH-like malate dehydrogenase from Ignicoccus islandicus displays dual substrate recognition, hidden allostery and a non-canonical tetrameric oligomeric organization.
J.Struct.Biol., 208, 2019
|
|
5J9I
 
 | |
6QNM
 
 | |
6OP6
 
 | | Structure of VIM-20 in the reduced state | | Descriptor: | Metallo-beta-lactamase VIM-20, SODIUM ION, ZINC ION | | Authors: | Page, R.C, Shurina, B.A, Montgomery, J.S, Orischak, M.G, Nix, J.C. | | Deposit date: | 2019-04-24 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A Single Salt Bridge in VIM-20 Increases Protein Stability and Antibiotic Resistance under Low-Zinc Conditions.
Mbio, 10, 2019
|
|
5IV4
 
 | | Crystal structure of the human soluble adenylyl cyclase in complex with the allosteric inhibitor LRE1 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-N~4~-cyclopropyl-N~4~-[(thiophen-2-yl)methyl]pyrimidine-2,4-diamine, ACETATE ION, ... | | Authors: | Kleinboelting, S, Steegborn, C. | | Deposit date: | 2016-03-18 | | Release date: | 2016-08-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of LRE1 as a specific and allosteric inhibitor of soluble adenylyl cyclase.
Nat.Chem.Biol., 12, 2016
|
|
6DHH
 
 | | RT XFEL structure of Photosystem II 400 microseconds after the second illumination at 2.2 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Chatterjee, R, Young, I.D, Fuller, F.D, Lassalle, L, Ibrahim, M, Gul, S, Fransson, T, Brewster, A.S, Alonso-Mori, R, Hussein, R, Zhang, M, Douthit, L, de Lichtenberg, C, Cheah, M.H, Shevela, D, Wersig, J, Seufert, I, Sokaras, D, Pastor, E, Weninger, C, Kroll, T, Sierra, R.G, Aller, P, Butryn, A, Orville, A.M, Liang, M, Batyuk, A, Koglin, J.E, Carbajo, S, Boutet, S, Moriarty, N.W, Holton, J.M, Dobbek, H, Adams, P.D, Bergmann, U, Sauter, N.K, Zouni, A, Messinger, J, Yano, J, Yachandra, V.K. | | Deposit date: | 2018-05-20 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of the intermediates of Kok's photosynthetic water oxidation clock.
Nature, 563, 2018
|
|
6ATX
 
 | |
1L2N
 
 | | Smt3 Solution Structure | | Descriptor: | Ubiquitin-like protein SMT3 | | Authors: | Sheng, W, Liao, X. | | Deposit date: | 2002-02-22 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a yeast ubiquitin-like protein Smt3: the role of structurally less defined sequences in protein-protein recognitions.
Protein Sci., 11, 2002
|
|
5F6Y
 
 | | Crystal structure of Ubc9 (K48/K49A/E54A) complexed with Fragment 2 (mercaptobenzoxazole) | | Descriptor: | 5-chloranyl-3~{H}-1,3-benzoxazole-2-thione, SUMO-conjugating enzyme UBC9 | | Authors: | Lountos, G.T, Hewitt, W.M, Zlotkowski, K, Dahlhauser, S, Saunders, L.B, Needle, D, Tropea, J.E, Zhan, C, Wei, G, Ma, B, Nussinov, R, Schneekloth, J.S.Jr, Waugh, D.S. | | Deposit date: | 2015-12-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Insights Into the Allosteric Inhibition of the SUMO E2 Enzyme Ubc9.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
