250L
 
 | | THE RESPONSE OF T4 LYSOZYME TO LARGE-TO-SMALL SUBSTITUTIONS WITHIN THE CORE AND ITS RELATION TO THE HYDROPHOBIC EFFECT | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, T4 LYSOZYME | | Authors: | Xu, J, Baase, W.A, Baldwin, E, Matthews, B.W. | | Deposit date: | 1997-10-22 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The response of T4 lysozyme to large-to-small substitutions within the core and its relation to the hydrophobic effect.
Protein Sci., 7, 1998
|
|
1F4V
 
 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY BOUND TO THE N-TERMINUS OF FLIM | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, FLAGELLAR MOTOR SWITCH PROTEIN, ... | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Henderson, R.K, King, D, Huang, L.S, Kustu, S, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-06-10 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of an activated response regulator bound to its target.
Nat.Struct.Biol., 8, 2001
|
|
1F6I
 
 | |
1F6J
 
 | |
1F6D
 
 | | THE STRUCTURE OF UDP-N-ACETYLGLUCOSAMINE 2-EPIMERASE FROM E. COLI. | | Descriptor: | CHLORIDE ION, SODIUM ION, UDP-N-ACETYLGLUCOSAMINE 2-EPIMERASE, ... | | Authors: | Campbell, R.E, Mosimann, S.C, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 2000-06-21 | | Release date: | 2000-12-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of UDP-N-acetylglucosamine 2-epimerase reveals homology to phosphoglycosyl transferases.
Biochemistry, 39, 2000
|
|
1FSF
 
 | | GLUCOSAMINE-6-PHOSPHATE DEAMINASE FROM E.COLI, T CONFORMER, AT 1.9A RESOLUTION | | Descriptor: | GLUCOSAMINE-6-PHOSPHATE DEAMINASE | | Authors: | Rudino-Pinera, E, Morales-Arrieta, S, Rojas-Trejo, S.P, Horjales, E. | | Deposit date: | 2000-09-08 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural flexibility, an essential component of the allosteric activation in Escherichia coli glucosamine-6-phosphate deaminase.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1F76
 
 | | ESCHERICHIA COLI DIHYDROOROTATE DEHYDROGENASE | | Descriptor: | Dihydroorotate dehydrogenase (quinone), FLAVIN MONONUCLEOTIDE, FORMIC ACID, ... | | Authors: | Norager, S, Jensen, K.F, Bjornberg, O, Larsen, S. | | Deposit date: | 2000-06-26 | | Release date: | 2002-10-16 | | Last modified: | 2014-03-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | E. coli Dihydroorotate Dehydrogenase Reveals Structural and Functional Distinction between different classes of
dihydroorotate dehydrogenases
Structure, 10, 2002
|
|
2AU7
 
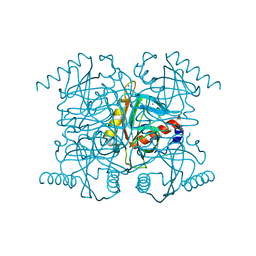 | | The R43Q active site variant of E.coli inorganic pyrophosphatase | | Descriptor: | CHLORIDE ION, Inorganic pyrophosphatase, MANGANESE (II) ION, ... | | Authors: | Samygina, V.R, Avaeva, S.M, Bartunik, H.D. | | Deposit date: | 2005-08-27 | | Release date: | 2006-08-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Reversible inhibition of Escherichia coli inorganic pyrophosphatase by fluoride: trapped catalytic intermediates in cryo-crystallographic studies
J.Mol.Biol., 366, 2007
|
|
1HQG
 
 | | CRYSTAL STRUCTURE OF THE H141C ARGINASE VARIANT COMPLEXED WITH PRODUCTS ORNITHINE AND UREA | | Descriptor: | ARGINASE 1, L-ornithine, MANGANESE (II) ION, ... | | Authors: | Cox, J.D, Cama, E, Colleluori, D.M, Ash, D.E, Christianson, D.W. | | Deposit date: | 2000-12-16 | | Release date: | 2001-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mechanistic and metabolic inferences from the binding of substrate analogues and products to arginase.
Biochemistry, 40, 2001
|
|
2R1H
 
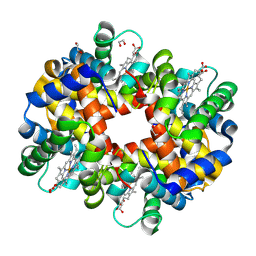 | | met-Trout IV hemoglobin at pH 6.3 | | Descriptor: | 1,2-ETHANEDIOL, Hemoglobin subunit alpha-4, Hemoglobin subunit beta-4, ... | | Authors: | Aranda IV, R, Worley, C.E, Richards, M.P, Phillips Jr, G.N. | | Deposit date: | 2007-08-22 | | Release date: | 2008-09-02 | | Last modified: | 2013-01-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of fish versus mammalian hemoglobins: Effect of the heme pocket environment on autooxidation and hemin loss.
Proteins, 75, 2008
|
|
2QRY
 
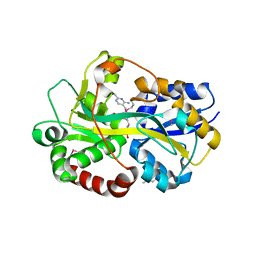 | | Periplasmic thiamin binding protein | | Descriptor: | THIAMIN PHOSPHATE, Thiamine-binding periplasmic protein | | Authors: | Ealick, S.E, Soriano, E.V. | | Deposit date: | 2007-07-30 | | Release date: | 2008-02-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Similarities between Thiamin-Binding Protein and Thiaminase-I Suggest a Common Ancestor
Biochemistry, 47, 2008
|
|
1VJM
 
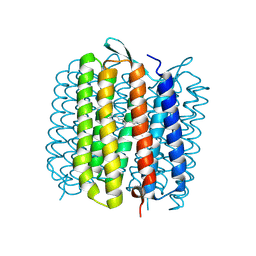 | | Deformation of helix C in the low-temperature L-intermediate of bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Edman, K, Royant, A, Larsson, G, Jacobson, F, Taylor, T, van der Spoel, D, Landau, E.M, Pebay-Peyroula, E, Neutze, R. | | Deposit date: | 2004-03-12 | | Release date: | 2004-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Deformation of helix C in the low temperature L-intermediate of bacteriorhodopsin.
J.Biol.Chem., 279, 2004
|
|
6Z4T
 
 | |
6Z4R
 
 | |
1HXD
 
 | | CRYSTAL STRUCTURE OF E. COLI BIOTIN REPRESSOR WITH BOUND BIOTIN | | Descriptor: | BIOTIN, BIRA BIFUNCTIONAL PROTEIN | | Authors: | Kwon, K, Streaker, E.D, Ruparelia, S, Beckett, D. | | Deposit date: | 2001-01-12 | | Release date: | 2001-05-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Corepressor-induced organization and assembly of the biotin repressor: a model for allosteric activation of a transcriptional regulator.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5DSF
 
 | | Crystal structure of the mercury-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, MERCURY (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-09-17 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.954 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
1HXQ
 
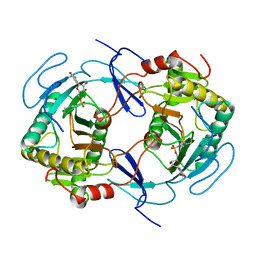 | | THE STRUCTURE OF NUCLEOTIDYLATED GALACTOSE-1-PHOSPHATE URIDYLYLTRANSFERASE FROM ESCHERICHIA COLI AT 1.86 ANGSTROMS RESOLUTION | | Descriptor: | FE (III) ION, HEXOSE-1-PHOSPHATE URIDYLYLTRANSFERASE, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Wedekind, J.E, Frey, P.A, Rayment, I. | | Deposit date: | 1996-06-16 | | Release date: | 1997-10-22 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The structure of nucleotidylated histidine-166 of galactose-1-phosphate uridylyltransferase provides insight into phosphoryl group transfer.
Biochemistry, 35, 1996
|
|
3UNA
 
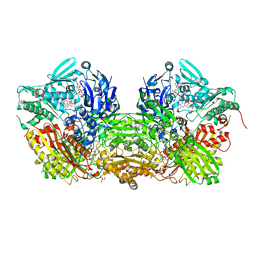 | | Crystal Structure of Bovine Milk Xanthine Dehydrogenase with NAD Bound | | Descriptor: | 2-HYDROXYBENZOIC ACID, CALCIUM ION, CARBONATE ION, ... | | Authors: | Eger, B.T, Okamoto, K, Nishino, T, Pai, E.F. | | Deposit date: | 2011-11-15 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein conformational gating of enzymatic activity in xanthine oxidoreductase.
J.Am.Chem.Soc., 134, 2012
|
|
8TJ3
 
 | |
4Z8W
 
 | |
2R96
 
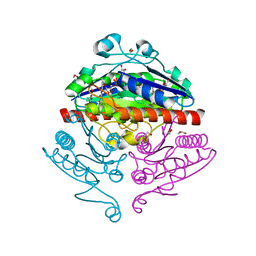 | | Crystal structure of E. coli WrbA in complex with FMN | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN MONONUCLEOTIDE, Flavoprotein WrbA | | Authors: | Kuta Smatanova, I, Wolfova, J, Brynda, J, Mesters, J.R, Grandori, R, Carey, J. | | Deposit date: | 2007-09-12 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural organization of WrbA in apo- and holoprotein crystals.
Biochim.Biophys.Acta, 1794, 2009
|
|
1DFO
 
 | | CRYSTAL STRUCTURE AT 2.4 ANGSTROM RESOLUTION OF E. COLI SERINE HYDROXYMETHYLTRANSFERASE IN COMPLEX WITH GLYCINE AND 5-FORMYL TETRAHYDROFOLATE | | Descriptor: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, SERINE HYDROXYMETHYLTRANSFERASE | | Authors: | Scarsdale, J.N, Radaev, S, Kazanina, G, Schirch, V, Wright, H.T. | | Deposit date: | 1999-11-20 | | Release date: | 1999-12-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure at 2.4 A resolution of E. coli serine hydroxymethyltransferase in complex with glycine substrate and 5-formyl tetrahydrofolate.
J.Mol.Biol., 296, 2000
|
|
4G1B
 
 | | X-ray structure of yeast flavohemoglobin in complex with econazole | | Descriptor: | 1-[(2S)-2-[(4-CHLOROBENZYL)OXY]-2-(2,4-DICHLOROPHENYL)ETHYL]-1H-IMIDAZOLE, FLAVIN-ADENINE DINUCLEOTIDE, Flavohemoglobin, ... | | Authors: | El Hammi, E, Warkentin, E, Demmer, U, Baciou, L, Ermler, U. | | Deposit date: | 2012-07-10 | | Release date: | 2012-11-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Active site analysis of yeast flavohemoglobin based on its structure with a small ligand or econazole.
Febs J., 279, 2012
|
|
3HZH
 
 | | Crystal structure of the CheX-CheY-BeF3-Mg+2 complex from Borrelia burgdorferi | | Descriptor: | Chemotaxis operon protein (CheX), Chemotaxis response regulator (CheY-3), MAGNESIUM ION | | Authors: | Pazy, Y, Silversmith, R.E, Guarinari, M, Zhao, R. | | Deposit date: | 2009-06-23 | | Release date: | 2010-02-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identical phosphatase mechanisms achieved through distinct modes of binding phosphoprotein substrate.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2R97
 
 | | Crystal structure of E. coli WrbA in complex with FMN | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavoprotein WrbA | | Authors: | Kuta Smatanova, I, Wolfova, J, Brynda, J, Mesters, J.R, Grandori, R, Carey, J. | | Deposit date: | 2007-09-12 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural organization of WrbA in apo- and holoprotein crystals.
Biochim.Biophys.Acta, 1794, 2009
|
|
