5ENF
 
 | | Crystal structure of the second bromodomain of Pleckstrin homology domain interacting protein (PHIP) in complex with fragment-4 N10142 (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 5-azanyl-2-(2-methylpropyl)-1,3-oxazole-4-carbonitrile, PH-interacting protein | | Authors: | Krojer, T, Talon, R, Collins, P, Bradley, A, Cox, O, Amin, J, Szykowska, A, Burgess-Brown, N, Spencer, J, Brennan, P, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-11-09 | | Release date: | 2016-04-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain.
Chem Sci, 7, 2016
|
|
2Q0N
 
 | | Structure of human p21 activating kinase 4 (PAK4) in complex with a consensus peptide | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Serine/threonine-protein kinase PAK 4, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Turnbull, A, Papagrigoriou, E, Pike, A.W, von Delft, F, Sundstrom, M, Edwards, A, Arrowsmith, C.H, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-22 | | Release date: | 2007-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of human p21 activating kinase 4 (PAK4) in complex with a consensus peptide.
To be Published
|
|
4O15
 
 | | The crystal structure of a mutant NAMPT (S165F) in complex with GNE-618 | | Descriptor: | 1,2-ETHANEDIOL, N-(4-{[3-(trifluoromethyl)phenyl]sulfonyl}benzyl)-2H-pyrazolo[3,4-b]pyridine-5-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Oh, A, Coons, M, Brillantes, B, Wang, W. | | Deposit date: | 2013-12-15 | | Release date: | 2014-10-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Resistance to Diverse Classes of NAMPT Inhibitors.
Plos One, 9, 2014
|
|
2WZZ
 
 | |
2PW2
 
 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide ELDKWKSL | | Descriptor: | 2F5 Fab' heavy chain, 2F5 Fab' light chain, peptide epitope | | Authors: | Bryson, S, Julien, J.-P, Hynes, R.C, Pai, E.F. | | Deposit date: | 2007-05-10 | | Release date: | 2007-05-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications.
J.Virol., 83, 2009
|
|
4AUD
 
 | |
7CHM
 
 | | Crystal structure of TTK kinase domain in complex with compound 8 | | Descriptor: | 4-(cyclohexylamino)-2-[(2-methoxy-4-morpholin-4-ylcarbonyl-phenyl)amino]-7H-pyrrolo[2,3-d]pyrimidine-5-carbonitrile, Dual specificity protein kinase TTK | | Authors: | Kim, H.L, Cho, H.Y, Park, Y.W, Lee, Y.H, Son, J.B, Ko, E.H, Choi, H.G, Kim, N.D. | | Deposit date: | 2020-07-06 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | X-ray Crystal Structure-Guided Design and Optimization of 7 H -Pyrrolo[2,3- d ]pyrimidine-5-carbonitrile Scaffold as a Potent and Orally Active Monopolar Spindle 1 Inhibitor.
J.Med.Chem., 64, 2021
|
|
1X36
 
 | | T=1 capsid of an amino-terminal deletion mutant of SeMV CP | | Descriptor: | CALCIUM ION, Coat protein | | Authors: | Sangita, V, Satheshkumar, P.S, Savithri, H.S, Murthy, M.R. | | Deposit date: | 2005-04-29 | | Release date: | 2005-10-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a mutant T=1 capsid of Sesbania mosaic virus: role of water molecules in capsid architecture and integrity.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7SCK
 
 | |
7SCJ
 
 | |
4AU7
 
 | | The structure of the Suv4-20h2 ternary complex with histone H4 | | Descriptor: | 1,2-ETHANEDIOL, HISTONE H4 PEPTIDE, HISTONE-LYSINE N-METHYLTRANSFERASE SUV420H2, ... | | Authors: | Southall, S.M, Cronin, N.B, Wilson, J.R. | | Deposit date: | 2012-05-14 | | Release date: | 2013-05-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | A Novel Route to Product Specificity in the Suv4-20 Family of Histone H4K20 Methyltransferases.
Nucleic Acids Res., 42, 2014
|
|
7FRT
 
 | | PanDDA analysis group deposition of ground-state model of PTP1B, using pre-clustering, cluster 1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-26 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
4FC0
 
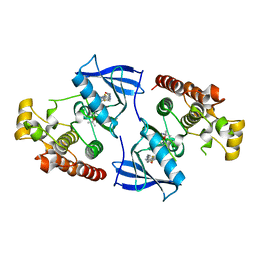 | | Crystal Structure of Human Kinase Domain of B-raf with a DFG-out Inhibitor | | Descriptor: | 2-chloro-3-[(2-cyanopropan-2-yl)oxy]-N-{5-[{2-[(cyclopropylcarbonyl)amino][1,3]thiazolo[5,4-b]pyridin-5-yl}(methyl)amino]-2-fluorophenyl}benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Yano, J.K, Aertgeerts, K. | | Deposit date: | 2012-05-23 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Design and synthesis of novel DFG-out RAF/vascular endothelial growth factor receptor 2 (VEGFR2) inhibitors: 3. Evaluation of 5-amino-linked thiazolo[5,4-d]pyrimidine and thiazolo[5,4-b]pyridine derivatives.
Bioorg.Med.Chem., 20, 2012
|
|
1M7Q
 
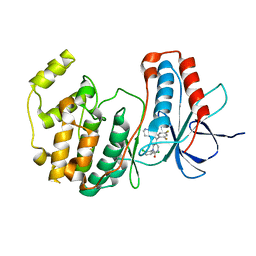 | | Crystal structure of p38 MAP kinase in complex with a dihydroquinazolinone inhibitor | | Descriptor: | 1-(2,6-DICHLOROPHENYL)-5-(2,4-DIFLUOROPHENYL)-7-PIPERAZIN-1-YL-3,4-DIHYDROQUINAZOLIN-2(1H)-ONE, Mitogen-activated protein kinase 14, SULFATE ION | | Authors: | Stelmach, J.E, Liu, L, Patel, S.B, Pivnichny, J.V, Scapin, G, Singh, S, Hop, C.E.C.A, Wang, Z, Cameron, P.M, Nichols, E.A, O'Keefe, S.J, O'Neill, E.A, Schmatz, D.M, Schwartz, C.D, Thompson, C.M, Zaller, D.M, Doherty, J.B. | | Deposit date: | 2002-07-22 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of potent, orally bioavailable dihydroquinazolinone inhibitors of p38 MAP kinase.
Bioorg.Med.Chem.Lett., 13, 2003
|
|
7FQX
 
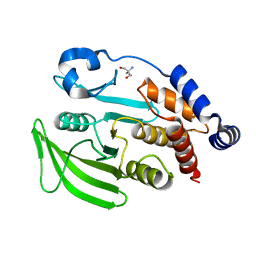 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with FMOPL000601a | | Descriptor: | 1-(3,4-dimethoxyphenyl)methanamine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-19 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
1X4M
 
 | | Solution structure of KH domain in Far upstream element binding protein 1 | | Descriptor: | Far upstream element binding protein 1 | | Authors: | He, F, Muto, Y, Inoue, M, Kigawa, T, Shirouzu, M, Terada, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-14 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of KH domain in Far upstream element binding protein 1
To be Published
|
|
4YON
 
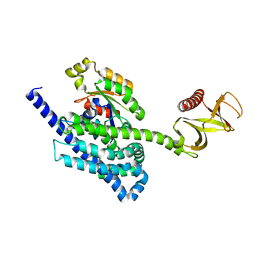 | | P-Rex1:Rac1 complex | | Descriptor: | Phosphatidylinositol 3,4,5-trisphosphate-dependent Rac exchanger 1 protein, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Lucato, C.M, Whisstock, J.C, Ellisdon, A.M. | | Deposit date: | 2015-03-11 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Phosphatidylinositol (3,4,5)-Trisphosphate-dependent Rac Exchanger 1Ras-related C3 Botulinum Toxin Substrate 1 (P-Rex1Rac1) Complex Reveals the Basis of Rac1 Activation in Breast Cancer Cells.
J.Biol.Chem., 290, 2015
|
|
7FRL
 
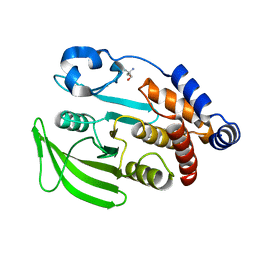 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with Z2856434917 | | Descriptor: | 1-(3,4-dimethoxyphenyl)methanamine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
3KLL
 
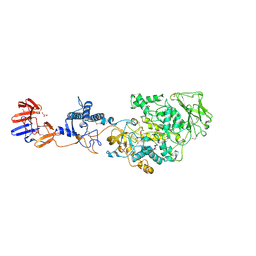 | | Crystal structure of Lactobacillus reuteri N-terminally truncated glucansucrase GTF180-maltose complex | | Descriptor: | CALCIUM ION, GLYCEROL, Glucansucrase, ... | | Authors: | Vujicic-Zagar, A, Pijning, T, Kralj, S, Eeuwema, W, Dijkhuizen, L, Dijkstra, B.W. | | Deposit date: | 2009-11-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a 117 kDa glucansucrase fragment provides insight into evolution and product specificity of GH70 enzymes
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7FRN
 
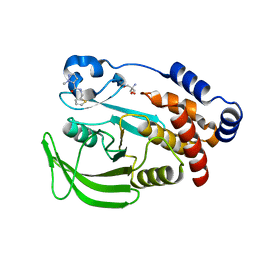 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with Z915492990 | | Descriptor: | 1-methyl-N-[(thiophen-2-yl)methyl]-1H-pyrazole-5-carboxamide, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
7FRJ
 
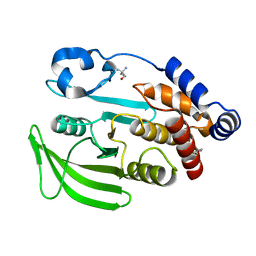 | | PanDDA analysis group deposition -- Crystal structure of PTP1B in complex with Z2856434770 | | Descriptor: | 1-(3-chlorophenyl)-N-methylmethanamine, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Mehlman, T, Biel, J, Azeem, S.M, Nelson, E.R, Hossain, S, Dunnett, L.E, Paterson, N.G, Douangamath, A, Talon, R, Axford, D, Orins, H, von Delft, F, Keedy, D.A. | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Room-temperature crystallography reveals altered binding of small-molecule fragments to PTP1B.
Elife, 12, 2023
|
|
6JAP
 
 | | Crystal structure of ABC transporter alpha-glycoside-binding mutant protein R356A in complex with sucrose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter, periplasmic substrate-binding protein, ... | | Authors: | Kanaujia, S.P, Chandravanshi, M, Gogoi, P. | | Deposit date: | 2019-01-24 | | Release date: | 2019-10-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural and thermodynamic correlation illuminates the selective transport mechanism of disaccharide alpha-glycosides through ABC transporter.
Febs J., 287, 2020
|
|
8UWA
 
 | | VH1-18 QxxV class antibody 09-1B12 bound to A/Perth/16/2009 H3N2 hemagglutinin | | Descriptor: | 09-1B12 heavy chain, 09-1B12 light chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maurer, D.P. | | Deposit date: | 2023-11-06 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.02 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
3KE7
 
 | |
1VPU
 
 | |
