3J2J
 
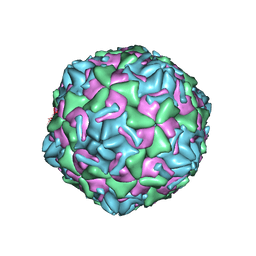 | | Empty coxsackievirus A9 capsid | | Descriptor: | Protein VP1, Protein VP2, Protein VP3 | | Authors: | Shakeel, S, Seitsonen, J.J.T, Kajander, T, Laurinmaki, P, Hyypia, T, Susi, P, Butcher, S.J. | | Deposit date: | 2012-10-04 | | Release date: | 2013-07-17 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.54 Å) | | Cite: | Structural and functional analysis of coxsackievirus A9 integrin {alpha}v{beta}6 binding and uncoating.
J.Virol., 87, 2013
|
|
3J9F
 
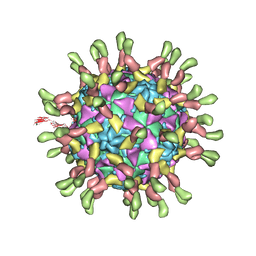 | | Poliovirus complexed with soluble, deglycosylated poliovirus receptor (Pvr) at 4 degrees C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Strauss, M, Filman, D.J, Belnap, D.M, Cheng, N, Noel, R.T, Hogle, J.M. | | Deposit date: | 2015-01-15 | | Release date: | 2015-02-11 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Nectin-Like Interactions between Poliovirus and Its Receptor Trigger Conformational Changes Associated with Cell Entry.
J.Virol., 89, 2015
|
|
3J6M
 
 | | Kinetic and Structural Analysis of Coxsackievirus B3 Receptor Interactions and Formation of the A-particle | | Descriptor: | Coxsackievirus and adenovirus receptor | | Authors: | Organtini, L.J, Makhov, A.M, Conway, J.F, Hafenstein, S, Carson, S.D. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-09 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Kinetic and structural analysis of coxsackievirus b3 receptor interactions and formation of the a-particle.
J.Virol., 88, 2014
|
|
3J6O
 
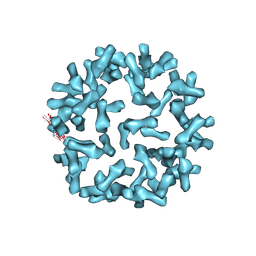 | | Kinetic and Structural Analysis of Coxsackievirus B3 Receptor Interactions and Formation of the A-particle | | Descriptor: | Coxsackie and adenovirus receptor | | Authors: | Organtini, L.J, Makhov, A.M, Conway, J.F, Hafenstein, S, Carson, S.D. | | Deposit date: | 2014-03-19 | | Release date: | 2014-04-09 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | Kinetic and structural analysis of coxsackievirus b3 receptor interactions and formation of the a-particle.
J.Virol., 88, 2014
|
|
3J3O
 
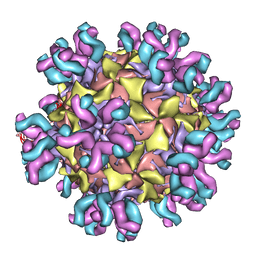 | | Conformational Shift of a Major Poliovirus Antigen Confirmed by Immuno-Cryogenic Electron Microscopy: 160S Poliovirus and C3-Fab Complex | | Descriptor: | C3 antibody, heavy chain, light chain, ... | | Authors: | Lin, J, Cheng, N, Hogle, J.M, Steven, A.C, Belnap, D.M. | | Deposit date: | 2013-04-10 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11.1 Å) | | Cite: | Conformational shift of a major poliovirus antigen confirmed by immuno-cryogenic electron microscopy.
J.Immunol., 191, 2013
|
|
6ADM
 
 | |
6ADL
 
 | |
6AJ2
 
 | | The structure of ICAM-5 triggered Enterovirus D68 virus A-particle | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AKS
 
 | | Cryo-EM structure of CVA10 mature virus | | Descriptor: | SPHINGOSINE, VP1, VP2, ... | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6AJ0
 
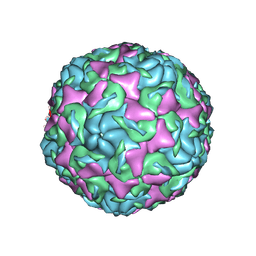 | | The structure of Enterovirus D68 mature virion | | Descriptor: | Capsid protein VP3, Capsid protein VP4, Viral protein 1, ... | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-25 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AKT
 
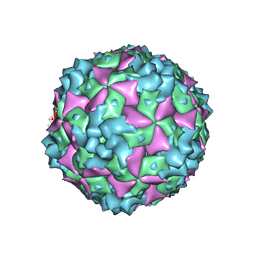 | | Cryo-EM structure of CVA10 A-particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6AJ9
 
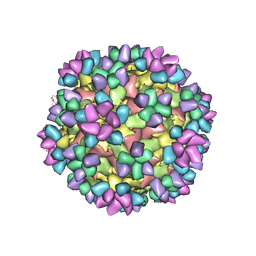 | | The structure of Enterovirus D68 mature virion in complex with Fab 15C5 and 11G1 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-27 | | Release date: | 2018-11-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AKU
 
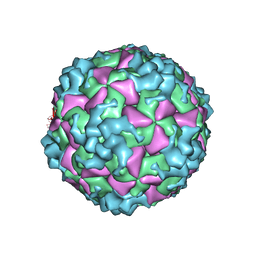 | | Cryo-EM structure of CVA10 empty particle | | Descriptor: | VP1, VP2, VP3 | | Authors: | Zhu, L, Sun, Y, Fan, J.Y, Zhu, B, Cao, L, Gao, Q, Zhang, Y.J, Liu, H.R, Rao, Z.H, Wang, X.X. | | Deposit date: | 2018-09-03 | | Release date: | 2019-01-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures of Coxsackievirus A10 unveil the molecular mechanisms of receptor binding and viral uncoating.
Nat Commun, 9, 2018
|
|
6AJ3
 
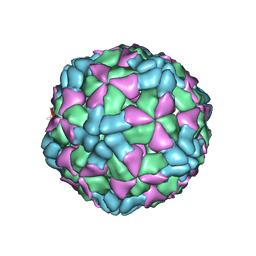 | | The structure of Enterovirus D68 procapsid | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-26 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
6AJ7
 
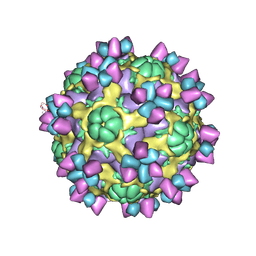 | | The structure of Enterovirus D68 mature virion in complex with Fab 15C5 | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Zheng, Q.B, Zhu, R, Xu, L.F, He, M.Z, Yan, X.D, Cheng, T, Li, S.W. | | Deposit date: | 2018-08-27 | | Release date: | 2018-11-07 | | Last modified: | 2022-03-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Atomic structures of enterovirus D68 in complex with two monoclonal antibodies define distinct mechanisms of viral neutralization
Nat Microbiol, 4, 2019
|
|
2R70
 
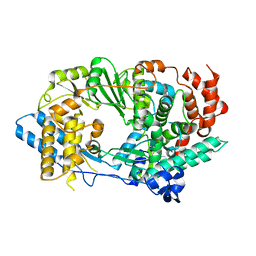 | | Crystal structure of infectious bursal disease virus VP1 polymerase, cocrystallized with an oligopeptide mimicking the VP3 C-terminus. | | Descriptor: | INFECTIOUS BURSAL VIRUS VP1 POLYMERASE | | Authors: | Garriga, D, Navarro, A, Querol-Audi, J, Abaitua, F, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2007-09-07 | | Release date: | 2007-11-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Activation mechanism of a noncanonical RNA-dependent RNA polymerase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PUS
 
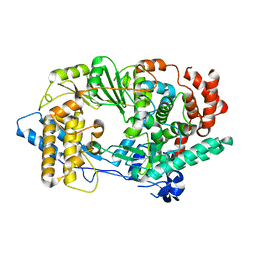 | | Unprecedented activation mechanism of a non-canonical RNA-dependent RNA polymerase | | Descriptor: | IBDV VP1 RNA-dependant RNA polymerase | | Authors: | Garriga, D, Navarro, A, Querol-Audi, J, Abaitua, F, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2007-05-09 | | Release date: | 2007-11-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Activation mechanism of a noncanonical RNA-dependent RNA polymerase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2R72
 
 | | Crystal structure of infectious bursal disease virus VP1 polymerase, incubated with Mg2+ ion. | | Descriptor: | INFECTIOUS BURSAL DISEASE VIRUS VP1 POLYMERASE, MAGNESIUM ION | | Authors: | Garriga, D, Navarro, A, Querol-Audi, J, Abaitua, F, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2007-09-07 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Activation mechanism of a noncanonical RNA-dependent RNA polymerase.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4NLO
 
 | | Poliovirus Polymerase - C290I Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
4NLU
 
 | | Poliovirus Polymerase - G289A Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
4NLV
 
 | | Poliovirus Polymerase - G289A/C290F Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
4NLW
 
 | | Poliovirus Polymerase - G289A/C290I Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
4NLT
 
 | | Poliovirus Polymerase - S291P Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
4NLY
 
 | | Poliovirus Polymerase - C290E Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
4NLR
 
 | | Poliovirus Polymerase - C290S Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
