1XI9
 
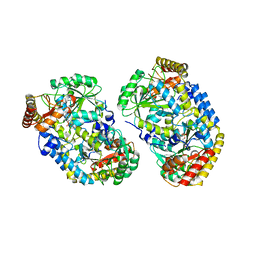 | | Alanine aminotransferase from Pyrococcus furiosus Pfu-1397077-001 | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, putative transaminase | | Authors: | Zhou, W, Tempel, W, Shah, A, Chen, L, Liu, Z.-J, Lee, D, Lin, D, Chang, S.-H, Brereton, P.S, Izumi, M, Jenney Jr, F.E, Lee, H.-S, Poole II, F.L, Shah, C, Sugar, F.J, Arendall III, W.B, Richardson, J.S, Richardson, D.C, Rose, J.P, Adams, M.W.W, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-09-21 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Alanine aminotransferase from Pyrococcus furiosus Pfu-1397077-001
To be published
|
|
1XIA
 
 | |
1XIB
 
 | |
1XIC
 
 | |
1XID
 
 | |
1XIE
 
 | |
1XIF
 
 | |
1XIG
 
 | |
1XIH
 
 | |
1XII
 
 | |
1XIJ
 
 | |
1XIK
 
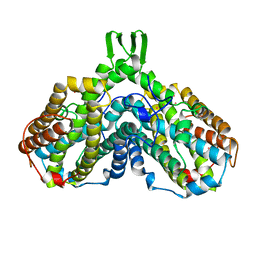 | | RIBONUCLEOSIDE-DIPHOSPHATE REDUCTASE 1 BETA CHAIN | | Descriptor: | FE (II) ION, MERCURY (II) ION, PROTEIN R2 OF RIBONUCLEOTIDE REDUCTASE | | Authors: | Logan, D.T, Su, X.-D, Aberg, A, Regnstrom, K, Hajdu, J, Eklund, H, Nordlund, P. | | Deposit date: | 1996-08-06 | | Release date: | 1997-03-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of reduced protein R2 of ribonucleotide reductase: the structural basis for oxygen activation at a dinuclear iron site.
Structure, 4, 1996
|
|
1XIL
 
 | | HYDROGEN BONDING IN HUMAN MANGANESE SUPEROXIDE DISMUTASE CONTAINING 3-FLUOROTYROSINE | | Descriptor: | MANGANESE (II) ION, Superoxide dismutase [Mn], mitochondrial | | Authors: | Ayala, I, Perry, J.J, Szczepanski, J, Cabelli, D.E, Tainer, J.A, Vala, M.T, Nick, H.S, Silverman, D.N. | | Deposit date: | 2004-09-21 | | Release date: | 2005-03-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Hydrogen bonding in human manganese superoxide dismutase containing 3-fluorotyrosine
Biophys.J., 89, 2005
|
|
1XIM
 
 | | ARGININE RESIDUES AS STABILIZING ELEMENTS IN PROTEINS | | Descriptor: | COBALT (II) ION, D-XYLOSE ISOMERASE, Xylitol | | Authors: | Mrabet, N.T, Van Denbroek, A, Van Den Brande, I, Stanssens, P, Laroche, Y, Lambeir, A.-M, Matthyssens, G, Jenkins, J, Chiadmi, M, Vantilbeurgh, H, Rey, F, Janin, J, Quax, W.J, Lasters, I, Demaeyer, M, Wodak, S.J. | | Deposit date: | 1991-05-29 | | Release date: | 1993-04-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Arginine residues as stabilizing elements in proteins.
Biochemistry, 31, 1992
|
|
1XIN
 
 | |
1XIO
 
 | | Anabaena sensory rhodopsin | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ANABAENA SENSORY RHODOPSIN, RETINAL | | Authors: | Vogeley, L, Luecke, H. | | Deposit date: | 2004-09-21 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Anabaena sensory rhodopsin: a photochromic color sensor at 2.0 A.
Science, 306, 2004
|
|
1XIP
 
 | |
1XIQ
 
 | |
1XIS
 
 | |
1XIU
 
 | | Crystal structure of the agonist-bound ligand-binding domain of Biomphalaria glabrata RXR | | Descriptor: | (9cis)-retinoic acid, Nuclear receptor coactivator 1, RXR-like protein | | Authors: | De Groot, A, De Rosny, E, Juillan-Binard, C, Ferrer, J.-L, Laudet, V, Pebay-Peroula, E, Fontecilla-Camps, J.-C, Borel, F. | | Deposit date: | 2004-09-22 | | Release date: | 2005-09-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Novel Tetrameric Complex of Agonist-bound Ligand-binding Domain of Biomphalaria glabrata Retinoid X Receptor.
J.Mol.Biol., 354, 2005
|
|
1XIV
 
 | | Plasmodium falciparum lactate dehydrogenase complexed with 2-({4-chloro-[hydroxy(methoxy)methyl]cyclohexyl}amino)ethane-1,1,2-triol | | Descriptor: | 2-({4-CHLORO-2-[HYDROXY(METHOXY)METHYL]CYCLOHEXYL}AMINO)ETHANE-1,1,2-TRIOL, GLYCEROL, L-lactate dehydrogenase | | Authors: | Conners, R, Cameron, A, Read, J.A, Schambach, F, Sessions, R.B, Brady, R.L. | | Deposit date: | 2004-09-22 | | Release date: | 2005-06-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mapping the binding site for gossypol-like inhibitors of Plasmodium
falciparum lactate dehydrogenase
MOL.BIOCHEM.PARASITOL., 142, 2005
|
|
1XIW
 
 | | Crystal structure of human CD3-e/d dimer in complex with a UCHT1 single-chain antibody fragment | | Descriptor: | T-cell surface glycoprotein CD3 delta chain, T-cell surface glycoprotein CD3 epsilon chain, immunoglobulin heavy chain variable region, ... | | Authors: | Arnett, K.L, Harrison, S.C, Wiley, D.C. | | Deposit date: | 2004-09-22 | | Release date: | 2004-11-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a human CD3-epsilon/delta dimer in complex with a UCHT1 single-chain antibody fragment.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1XIX
 
 | |
1XIY
 
 | | Crystal Structure of Plasmodium falciparum antioxidant protein (1-Cys peroxiredoxin) | | Descriptor: | peroxiredoxin | | Authors: | Sarma, G.N, Fischer, M, Nickel, C, Becker, K, Karplus, P.A. | | Deposit date: | 2004-09-22 | | Release date: | 2005-02-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a novel Plasmodium falciparum 1-Cys peroxiredoxin.
J.Mol.Biol., 346, 2005
|
|
1XIZ
 
 | | Structural Genomics, The crystal structure of domain IIA of putative phosphotransferase system specific for mannitol/fructose from Salmonella typhimurium | | Descriptor: | putative phosphotransferase system mannitol/fructose-specific IIA domain | | Authors: | Zhang, R, Joachimiak, G, Otwinowski, Z, Collart, F, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-09-22 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of domain IIA of putative phosphotransferase system specific for mannitol/fructose from Salmonella typhimurium
To be Published
|
|
