5Z73
 
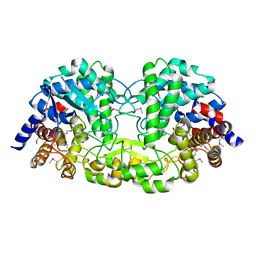 | | Crystal structure of alkaline/neutral invertase InvB from Anabaena sp. PCC 7120 | | Descriptor: | Alr0819 protein, GLYCEROL | | Authors: | Xie, J, Hu, H.X, Cai, K, Yang, F, Jiang, Y.L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2018-01-27 | | Release date: | 2018-05-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and enzymatic analyses of Anabaena heterocyst-specific alkaline invertase InvB.
FEBS Lett., 592, 2018
|
|
5IL2
 
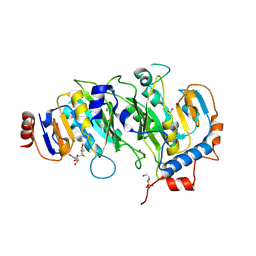 | | Crystal structure of SAH-bound METTL3-METTL14 complex | | Descriptor: | 1,2-ETHANEDIOL, METTL14, METTL3, ... | | Authors: | Wang, X, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-03-04 | | Release date: | 2016-05-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.606 Å) | | Cite: | Structural basis of N6-adenosine methylation by the METTL3-METTL14 complex
Nature, 534, 2016
|
|
7VRU
 
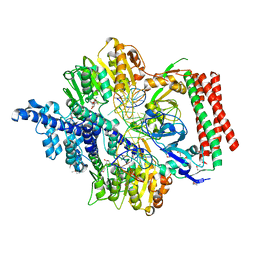 | | Crystal structure of PacII_M1M2S-DNA-SAH complex | | Descriptor: | DNA (25-mer), S-ADENOSYL-L-HOMOCYSTEINE, Site-specific DNA recognition subunit, ... | | Authors: | Zhu, J, Gao, P. | | Deposit date: | 2021-10-25 | | Release date: | 2022-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular insights into DNA recognition and methylation by non-canonical type I restriction-modification systems.
Nat Commun, 13, 2022
|
|
6I71
 
 | |
6E33
 
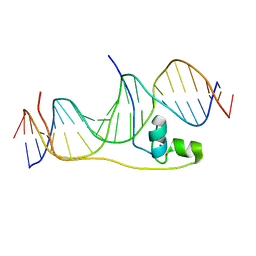 | | Crystal Structure of Pho7-DNA complex | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*TP*GP*AP*AP*TP*GP*TP*CP*CP*GP*AP*AP*GP*GP*AP*T)-3'), DNA (5'-D(*TP*CP*CP*TP*TP*CP*GP*GP*AP*CP*AP*TP*TP*CP*AP*AP*AP*TP*CP*A)-3'), Uncharacterized transcriptional regulatory protein C27B12.11c, ... | | Authors: | Garg, A, Goldgur, Y, Shuman, S. | | Deposit date: | 2018-07-13 | | Release date: | 2018-10-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Distinctive structural basis for DNA recognition by the fission yeast Zn2Cys6 transcription factor Pho7 and its role in phosphate homeostasis.
Nucleic Acids Res., 46, 2018
|
|
6AYQ
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with methylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-AMINO-5H-PYRROLO[3,2-D]PYRIMIDIN-7-YL)METHYL]-4-[(METHYLSULFANYL)METHYL]PYRROLIDIN-3-OL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, GLYCEROL | | Authors: | Cameron, S.A, Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AYT
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with pyrazinylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(pyrazin-2-ylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
2JJQ
 
 | | The crystal structure of Pyrococcus abyssi tRNA (uracil-54, C5)- methyltransferase in complex with S-adenosyl-L-homocysteine | | Descriptor: | GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, UNCHARACTERIZED RNA METHYLTRANSFERASE PYRAB10780 | | Authors: | Walbott, H, Leulliot, N, Grosjean, H, Golinelli-Pimpaneau, B. | | Deposit date: | 2008-04-17 | | Release date: | 2008-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Crystal Structure of Pyrococcus Abyssi tRNA (Uracil-54, C5)-Methyltransferase Provides Insights Into its tRNA Specificity.
Nucleic Acids Res., 36, 2008
|
|
2Q4S
 
 | | Ensemble refinement of the protein crystal structure of cysteine dioxygenase type I from Mus musculus Mm.241056 | | Descriptor: | 1,2-ETHANEDIOL, Cysteine dioxygenase type 1, NICKEL (II) ION | | Authors: | Levin, E.J, Kondrashov, D.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ensemble refinement of protein crystal structures: validation and application.
Structure, 15, 2007
|
|
3P9K
 
 | |
4EVI
 
 | | Crystal Structure Analysis of Coniferyl Alcohol 9-O-Methyltransferase from Linum Nodiflorum in Complex with Coniferyl Alcohol 9-Methyl Ether and S -Adenosyl-L-Homocysteine | | Descriptor: | 2-methoxy-4-[(1E)-3-methoxyprop-1-en-1-yl]phenol, 4-[(1E)-3-hydroxyprop-1-en-1-yl]-2-methoxyphenol, Coniferyl alcohol 9-O-methyltransferase, ... | | Authors: | Wolters, S, Heine, A, Petersen, M. | | Deposit date: | 2012-04-26 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Structural analysis of coniferyl alcohol 9-O-methyltransferase from Linum nodiflorum reveals a novel active-site environment.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3TLJ
 
 | | Crystal structure of Trm14 from Pyrococcus furiosus in complex with S-adenosyl-L-homocysteine | | Descriptor: | ACETATE ION, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine N2-)-methyltransferase Trm14 | | Authors: | Fislage, M, Roovers, M, Tuszynska, I, Bujnicki, J.M, Droogmans, L, Versees, W. | | Deposit date: | 2011-08-30 | | Release date: | 2012-03-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the tRNA:m2G6 methyltransferase Trm14/TrmN from two domains of life.
Nucleic Acids Res., 40, 2012
|
|
1ODM
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ANAEROBIC AC-VINYLGLYCINE FE COMPLEX) | | Descriptor: | DELTA-(L-ALPHA-AMINOADIPOYL)-L-CYSTEINYL-D-VINYLGLYCINE, FE (II) ION, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Elkins, J.M, Rutledge, P.J, Burzlaff, N.I, Clifton, I.J, Adlington, R.M, Roach, P.L, Baldwin, J.E. | | Deposit date: | 2003-02-19 | | Release date: | 2003-06-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystallographic Studies on the Reaction of Isopenicillin N Synthase with an Unsaturated Substrate Analogue
Org.Biomol.Chem., 1, 2003
|
|
6AYO
 
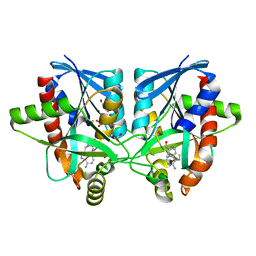 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with 5'-deoxy-5'-Propyl-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-propylpyrrolidin-3-ol, 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
1BC5
 
 | |
6AYR
 
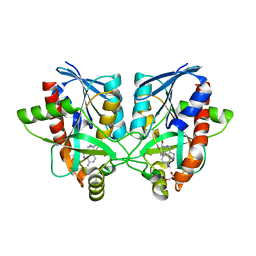 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with butylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(butylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
1BK0
 
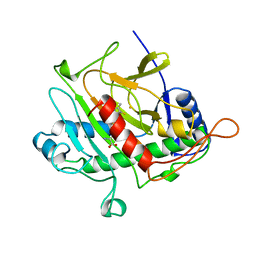 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ACV-FE COMPLEX) | | Descriptor: | FE (III) ION, ISOPENICILLIN N SYNTHASE, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, ... | | Authors: | Roach, P.L, Clifton, I.J, Hensgens, C.M.H, Shibata, N, Schofield, C.J, Hajdu, J, Baldwin, J.E. | | Deposit date: | 1998-07-14 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation.
Nature, 387, 1997
|
|
1BLZ
 
 | | ISOPENICILLIN N SYNTHASE FROM ASPERGILLUS NIDULANS (ACV-FE-NO COMPLEX) | | Descriptor: | FE (III) ION, ISOPENICILLIN N SYNTHASE, L-D-(A-AMINOADIPOYL)-L-CYSTEINYL-D-VALINE, ... | | Authors: | Roach, P.L, Clifton, I.J, Hensgens, C.M.H, Shibata, N, Schofield, C.J, Hajdu, J, Baldwin, J.E. | | Deposit date: | 1998-07-22 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of isopenicillin N synthase complexed with substrate and the mechanism of penicillin formation.
Nature, 387, 1997
|
|
6AYM
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) | | Descriptor: | 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
6AYS
 
 | | Crystal structure of Campylobacter jejuni 5'-methylthioadenosine/S-adenosyl homocysteine nucleosidase (MTAN) complexed with hexylthio-DADMe-Immucillin-A | | Descriptor: | (3R,4S)-1-[(4-amino-5H-pyrrolo[3,2-d]pyrimidin-7-yl)methyl]-4-[(hexylsulfanyl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-09-08 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Transition-State Analogues of Campylobacter jejuni 5'-Methylthioadenosine Nucleosidase.
ACS Chem. Biol., 13, 2018
|
|
1XCL
 
 | | Guanidinoacetate methyltransferase containing S-adenosylhomocysteine and guanidine | | Descriptor: | GUANIDINE, Guanidinoacetate N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Komoto, J, Yamada, T, Takata, Y, Takusagawa, F. | | Deposit date: | 2004-09-02 | | Release date: | 2004-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic mechanism of guanidinoacetate methyltransferase: crystal structures of guanidinoacetate methyltransferase ternary complexes.
Biochemistry, 43, 2004
|
|
1BVR
 
 | | M.TB. ENOYL-ACP REDUCTASE (INHA) IN COMPLEX WITH NAD+ AND C16-FATTY-ACYL-SUBSTRATE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (ENOYL-ACYL CARRIER PROTEIN (ACP) REDUCTASE), TRANS-2-HEXADECENOYL-(N-ACETYL-CYSTEAMINE)-THIOESTER | | Authors: | Rozwarski, D.A, Vilcheze, C, Sugantino, M, Bittman, R, Jacobs, W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 1998-09-17 | | Release date: | 1999-09-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Mycobacterium tuberculosis enoyl-ACP reductase, InhA, in complex with NAD+ and a C16 fatty acyl substrate.
J.Biol.Chem., 274, 1999
|
|
6I72
 
 | |
1M93
 
 | |
4E2X
 
 | | X-ray Structure of the Y222F mutant of TcaB9, a C-3'-Methyltransferase, in Complex with S-Adenosyl-L-Homocysteine and dTDP | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Bruender, N.A, Holden, H.M. | | Deposit date: | 2012-03-09 | | Release date: | 2012-04-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Probing the catalytic mechanism of a C-3'-methyltransferase involved in the biosynthesis of D-tetronitrose.
Protein Sci., 21, 2012
|
|
