1VM0
 
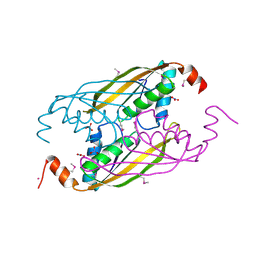 | | X-RAY STRUCTURE OF GENE PRODUCT FROM ARABIDOPSIS THALIANA AT2G34160 | | Descriptor: | NITRATE ION, POTASSIUM ION, unknown protein | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bitto, E, Bingman, C.A, Allard, S.T.M, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-08-24 | | Release date: | 2004-08-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-RAY STRUCTURE OF GENE PRODUCT FROM ARABIDOPSIS THALIANA AT2G34160
To be published
|
|
1VM1
 
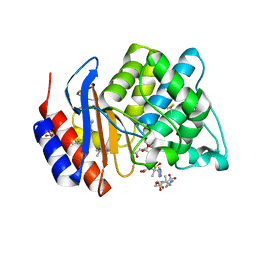 | | STRUCTURE OF SHV-1 BETA-LACTAMASE INHIBITED BY TAZOBACTAM | | Descriptor: | ACRYLIC ACID, BETA-LACTAMASE SHV-1, CYCLOHEXYL-HEXYL-BETA-D-MALTOSIDE, ... | | Authors: | Kuzin, A.P, Nukaga, M, Nukaga, Y, Hujer, A, Bonomo, R.A, Knox, J.R. | | Deposit date: | 2004-08-27 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Inhibition of the SHV-1 beta-lactamase by sulfones: crystallographic observation of two reaction intermediates with tazobactam.
Biochemistry, 40, 2001
|
|
1VM2
 
 | |
1VM3
 
 | |
1VM4
 
 | |
1VM5
 
 | |
1VM6
 
 | |
1VM7
 
 | |
1VM8
 
 | |
1VM9
 
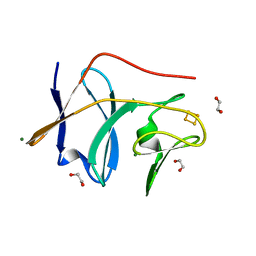 | | The X-ray Structure of the cys84ala cys85ala double mutant of the [2Fe-2S] Ferredoxin subunit of Toluene-4-Monooxygenase from Pseudomonas Mendocina KR1 | | Descriptor: | 1,2-ETHANEDIOL, FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION, ... | | Authors: | Wesenberg, G.E, Smith, D.W, Phillips Jr, G.N, Bingman, C.A, Allard, S.T.M, Moe, L.A, Fox, B.G, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-09-13 | | Release date: | 2004-09-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of T4moC, the Rieske-type ferredoxin component of toluene 4-monooxygenase.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1VMA
 
 | |
1VMB
 
 | |
1VMC
 
 | | STROMA CELL-DERIVED FACTOR-1ALPHA (SDF-1ALPHA) | | Descriptor: | Stromal cell-derived factor 1 | | Authors: | Gozansky, E.K, Clore, G.M. | | Deposit date: | 2004-09-20 | | Release date: | 2005-03-01 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Mapping the binding of the N-terminal extracellular tail of the CXCR4 receptor to stromal cell-derived factor-1alpha.
J.Mol.Biol., 345, 2005
|
|
1VMD
 
 | |
1VME
 
 | |
1VMF
 
 | |
1VMG
 
 | |
1VMH
 
 | |
1VMI
 
 | |
1VMJ
 
 | |
1VMK
 
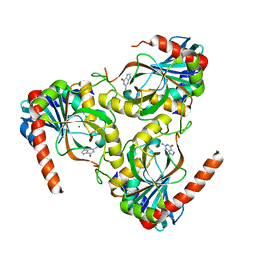 | |
1VMO
 
 | | CRYSTAL STRUCTURE OF VITELLINE MEMBRANE OUTER LAYER PROTEIN I (VMO-I): A FOLDING MOTIF WITH HOMOLOGOUS GREEK KEY STRUCTURES RELATED BY AN INTERNAL THREE-FOLD SYMMETRY | | Descriptor: | VITELLINE MEMBRANE OUTER LAYER PROTEIN I | | Authors: | Shimizu, T, Vassylyev, D.G, Kido, S, Doi, Y, Morikawa, K. | | Deposit date: | 1994-01-06 | | Release date: | 1994-05-31 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of vitelline membrane outer layer protein I (VMO-I): a folding motif with homologous Greek key structures related by an internal three-fold symmetry.
EMBO J., 13, 1994
|
|
1VMP
 
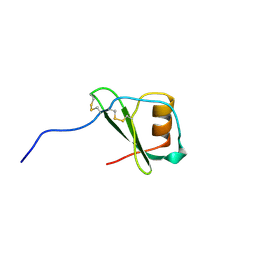 | | STRUCTURE OF THE ANTI-HIV CHEMOKINE VMIP-II | | Descriptor: | PROTEIN (ANTI-HIV CHEMOKINE MIP VII) | | Authors: | Liwang, A.C, Wang, Z.-X, Sun, Y, Peiper, S.C, Liwang, P.J. | | Deposit date: | 1999-03-25 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the anti-HIV chemokine vMIP-II.
Protein Sci., 8, 1999
|
|
1VNA
 
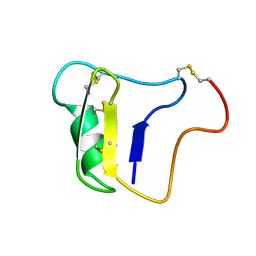 | |
1VNB
 
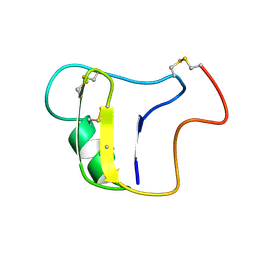 | |
