7W7I
 
 | |
3SQB
 
 | | Structure of the major type 1 pilus subunit FimA bound to the FimC chaperone | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chaperone protein fimC, ... | | Authors: | Scharer, M.A, Eidam, O, Grutter, M.G, Glockshuber, R, Capitani, G. | | Deposit date: | 2011-07-05 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Quality control of disulfide bond formation in pilus subunits by the chaperone FimC.
Nat.Chem.Biol., 8, 2012
|
|
5UVR
 
 | |
3PF2
 
 | |
5LP9
 
 | | FimA wt from S. flexneri | | Descriptor: | Major type 1 subunit fimbrin (Pilin) | | Authors: | Zyla, D, Capitani, G, Prota, A, Glockshuber, R. | | Deposit date: | 2016-08-12 | | Release date: | 2017-12-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (0.88626635 Å) | | Cite: | Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria.
J.Biol.Chem., 2019
|
|
7F7Y
 
 | |
8GR6
 
 | | Crystal Structure of pilus-specific Sortase C from Streptococcus sanguinis | | Descriptor: | 1,2-ETHANEDIOL, SODIUM ION, Sortase-like protein, ... | | Authors: | Yadav, S, Parijat, P, Krishnan, V. | | Deposit date: | 2022-09-01 | | Release date: | 2023-06-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal structure of the pilus-specific sortase from early colonizing oral Streptococcus sanguinis captures an active open-lid conformation.
Int.J.Biol.Macromol., 243, 2023
|
|
2OPE
 
 | | Crystal structure of the Neisseria meningitidis minor Type IV pilin, PilX, in space group P43 | | Descriptor: | PilX | | Authors: | Dyer, D.H, Helaine, S, Pelicic, V, Forest, K.T. | | Deposit date: | 2007-01-29 | | Release date: | 2007-10-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2OPD
 
 | | Structure of the Neisseria meningitidis minor Type IV pilin, PilX | | Descriptor: | PilX | | Authors: | Dyer, D.H, Helaine, S, Pelicic, V, Forest, K.T. | | Deposit date: | 2007-01-29 | | Release date: | 2007-10-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 3D structure/function analysis of PilX reveals how minor pilins can modulate the virulence properties of type IV pili.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1N0L
 
 | | Crystal structure of the PapD chaperone (C-terminally 6x histidine-tagged) bound to the PapE pilus subunit (N-terminal-deleted) from uropathogenic E. coli | | Descriptor: | Chaperone protein PapD, mature Fimbrial protein PapE | | Authors: | Sauer, F.G, Pinkner, J.S, Waksman, G, Hultgren, S.J. | | Deposit date: | 2002-10-14 | | Release date: | 2002-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation
Cell(Cambridge,Mass.), 111, 2002
|
|
5HBB
 
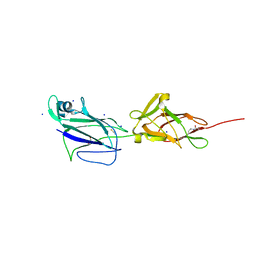 | | Crystal structure of shaft pilin spaA from Lactobacillus rhamnosus GG - E139A mutant | | Descriptor: | 1,2-ETHANEDIOL, Cell surface protein SpaA, SODIUM ION, ... | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2015-12-31 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
5HTS
 
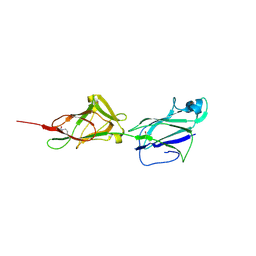 | | Crystal structure of shaft pilin spaA from Lactobacillus rhamnosus GG - D295N mutant | | Descriptor: | Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2016-01-27 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
4HHX
 
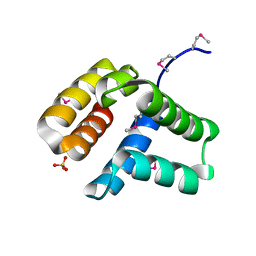 | | Structure of cytoplasmic domain of TCPE from Vibrio cholerae | | Descriptor: | SULFATE ION, Toxin coregulated pilus biosynthesis protein E | | Authors: | Kolappan, S, Craig, L. | | Deposit date: | 2012-10-10 | | Release date: | 2013-04-03 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure of the cytoplasmic domain of TcpE, the inner membrane core protein required for assembly of the Vibrio cholerae toxin-coregulated pilus.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HSS
 
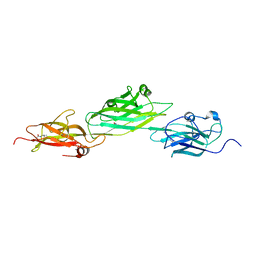 | |
4HSQ
 
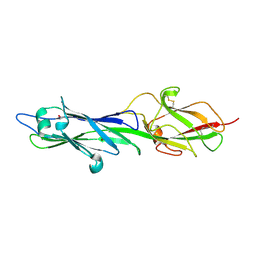 | |
5HDL
 
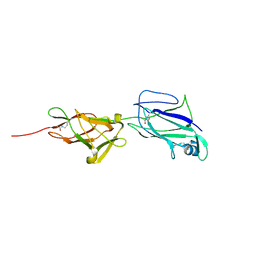 | | Crystal structure of shaft pilin spaA from Lactobacillus rhamnosus GG - E269A mutant | | Descriptor: | Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2016-01-05 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
5J4M
 
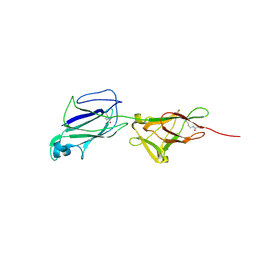 | | Crystal structure of shaft pilin SpaA from Lactobacillus rhamnosus GG - E269A/D295N double mutant | | Descriptor: | Cell surface protein SpaA | | Authors: | Chaurasia, P, Pratap, S, von Ossowski, I, Palva, A, Krishnan, V. | | Deposit date: | 2016-04-01 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | New insights about pilus formation in gut-adapted Lactobacillus rhamnosus GG from the crystal structure of the SpaA backbone-pilin subunit
Sci Rep, 6, 2016
|
|
2WMP
 
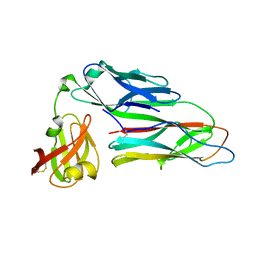 | | Structure of the E. coli chaperone PapD in complex with the pilin domain of the PapGII adhesin | | Descriptor: | CHAPERONE PROTEIN PAPD, PAPG PROTEIN | | Authors: | Ford, B.A, Verger, D, Elam, J.S, Dodson, K.W, Pinkner, J.S, Hultgren, S.J. | | Deposit date: | 2009-07-02 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Papd-Papgii Pilin Complex Reveals an Open and Flexible P5 Pocket.
J.Bacteriol., 194, 2012
|
|
7VCR
 
 | |
7W6B
 
 | |
7VCN
 
 | |
6XXE
 
 | |
6XXD
 
 | |
3G66
 
 | | The crystal structure of Streptococcus pneumoniae Sortase C provides novel insights into catalysis as well as pilin substrate specificity | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Sortase C | | Authors: | Neiers, F, Madhurantakam, C, Falker, S, Manzano, C, Dessen, A, Normark, S, Henriques-Normark, B, Achour, A. | | Deposit date: | 2009-02-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Two crystal structures of pneumococcal pilus sortase C provide novel insights into catalysis and substrate specificity.
J.Mol.Biol., 393, 2009
|
|
3G69
 
 | | The crystal structure of Streptococcus pneumoniae Sortase C provides novel insights into catalysis as well as pilin substrate specificity | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, SULFATE ION, Sortase C | | Authors: | Neiers, F, Madhurantakam, C, Falker, S, Manzano, C, Dessen, A, Normark, S, Henriques-Normark, B, Achour, A. | | Deposit date: | 2009-02-06 | | Release date: | 2009-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two crystal structures of pneumococcal pilus sortase C provide novel insights into catalysis and substrate specificity.
J.Mol.Biol., 393, 2009
|
|
