8A07
 
 | |
8A0T
 
 | | MTH1 in complex with TH012532 | | Descriptor: | 5-(phenylmethyl)pyrimidine-2,4-diol, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Scaletti, E, Stenmark, P. | | Deposit date: | 2022-05-30 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MTH1 in complex with TH012532
To Be Published
|
|
8A34
 
 | | MTH1 in complex with TH013071 | | Descriptor: | 5-(2-phenylphenyl)-1H-pyrimidine-2,4-dione, 7,8-dihydro-8-oxoguanine triphosphatase, ACETATE ION, ... | | Authors: | Liu, H, Scaletti, E, Stenmark, P. | | Deposit date: | 2022-06-07 | | Release date: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MTH1 in complex with TH013071
To Be Published
|
|
1G0S
 
 | |
1GA7
 
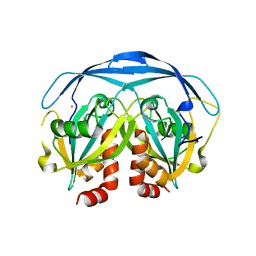 | | CRYSTAL STRUCTURE OF THE ADP-RIBOSE PYROPHOSPHATASE IN COMPLEX WITH GD+3 | | Descriptor: | GADOLINIUM ION, HYPOTHETICAL 23.7 KDA PROTEIN IN ICC-TOLC INTERGENIC REGION | | Authors: | Gabelli, S.B, Bianchet, M.A, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2000-11-29 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | The structure of ADP-ribose pyrophosphatase reveals the structural basis for the versatility of the Nudix family.
Nat.Struct.Biol., 8, 2001
|
|
4ICK
 
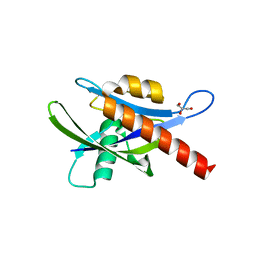 | | Crystal structure of human AP4A hydrolase E58A mutant | | Descriptor: | Bis(5'-nucleosyl)-tetraphosphatase [asymmetrical], GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ge, H, Chen, X. | | Deposit date: | 2012-12-10 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of wild-type and mutant human Ap4A hydrolase.
Biochem.Biophys.Res.Commun., 432, 2013
|
|
4IJX
 
 | |
4ILQ
 
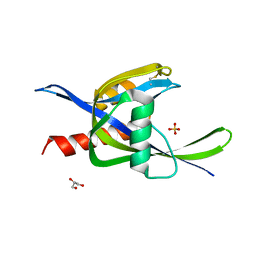 | | 2.60A resolution structure of CT771 from Chlamydia trachomatis | | Descriptor: | CT771, GLYCEROL, SULFATE ION | | Authors: | Barta, M.L, Lovell, S, Battaile, K.P, Hefty, P.S. | | Deposit date: | 2012-12-31 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chlamydia trachomatis CT771 (nudH) Is an Asymmetric Ap4A Hydrolase.
Biochemistry, 53, 2014
|
|
1G9Q
 
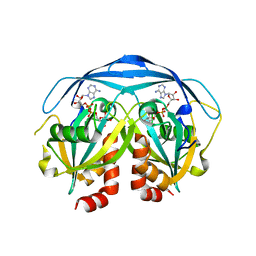 | | COMPLEX STRUCTURE OF THE ADPR-ASE AND ITS SUBSTRATE ADP-RIBOSE | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, HYPOTHETICAL 23.7 KDA PROTEIN IN ICC-TOLC INTERGENIC REGION | | Authors: | Gabelli, S.B, Bianchet, M.A, Bessman, M.J, Amzel, L.M. | | Deposit date: | 2000-11-27 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of ADP-ribose pyrophosphatase reveals the structural basis for the versatility of the Nudix family.
Nat.Struct.Biol., 8, 2001
|
|
1F3Y
 
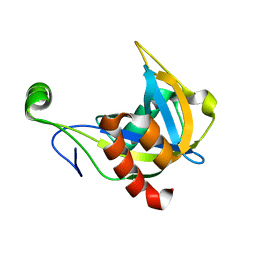 | | SOLUTION STRUCTURE OF THE NUDIX ENZYME DIADENOSINE TETRAPHOSPHATE HYDROLASE FROM LUPINUS ANGUSTIFOLIUS L. | | Descriptor: | DIADENOSINE 5',5'''-P1,P4-TETRAPHOSPHATE HYDROLASE | | Authors: | Swarbrick, J.D, Bashtannyk, T, Maksel, D, Zhang, X.R, Blackburn, G.M, Gayler, K.R, Gooley, P.R. | | Deposit date: | 2000-06-06 | | Release date: | 2001-06-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The three-dimensional structure of the Nudix enzyme diadenosine tetraphosphate hydrolase from Lupinus angustifolius L.
J.Mol.Biol., 302, 2000
|
|
1HX3
 
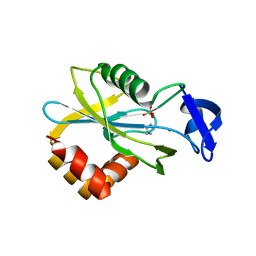 | | CRYSTAL STRUCTURE OF E.COLI ISOPENTENYL DIPHOSPHATE:DIMETHYLALLYL DIPHOSPHATE ISOMERASE | | Descriptor: | IMIDAZOLE, ISOPENTENYL DIPHOSPHATE DELTA-ISOMERASE, MANGANESE (II) ION, ... | | Authors: | Durbecq, V, Sainz, G, Oudjama, Y, Clantin, B, Bompard-Gilles, C, Tricot, C, Caillet, J, Stalon, V, Droogmans, L, Villeret, V. | | Deposit date: | 2001-01-11 | | Release date: | 2001-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of isopentenyl diphosphate:dimethylallyl diphosphate isomerase.
Embo J., 20, 2001
|
|
7R0D
 
 | |
7SEZ
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with m7GDP | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DNA repair NTP-phosphohydrolase, SODIUM ION | | Authors: | Peters, J.K, Tibble, R.W, Warminski, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.70001245 Å) | | Cite: | Structure of the poxvirus decapping enzyme D9 reveals its mechanism of cap recognition and catalysis.
Structure, 30, 2022
|
|
7SF0
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with trinucleotide substrate | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, DNA repair NTP-phosphohydrolase, MAGNESIUM ION, ... | | Authors: | Peters, J.K, Tibble, R.W, Warminski, M, Jemielity, J, Gross, J.D. | | Deposit date: | 2021-10-02 | | Release date: | 2022-03-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95000446 Å) | | Cite: | Structure of the poxvirus decapping enzyme D9 reveals its mechanism of cap recognition and catalysis.
Structure, 30, 2022
|
|
2RRK
 
 | |
1X84
 
 | | IPP isomerase (wt) reacted with (S)-bromohydrine of IPP | | Descriptor: | (S)-4-BROMO-3-HYDROXY-3-METHYLBUTYL DIPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, MAGNESIUM ION, ... | | Authors: | Wouters, J, Oldfield, E. | | Deposit date: | 2004-08-17 | | Release date: | 2005-01-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A Crystallographic Investigation of Phosphoantigen Binding to Isopentenyl Pyrophosphate/Dimethylallyl Pyrophosphate Isomerase
J.Am.Chem.Soc., 127, 2005
|
|
1X83
 
 | | Y104F IPP isomerase reacted with (S)-bromohydrine of IPP | | Descriptor: | (S)-4-BROMO-3-HYDROXY-3-METHYLBUTYL DIPHOSPHATE, Isopentenyl-diphosphate delta-isomerase, MAGNESIUM ION, ... | | Authors: | Wouters, J, Oldfield, E. | | Deposit date: | 2004-08-17 | | Release date: | 2005-01-25 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Crystallographic Investigation of Phosphoantigen Binding to Isopentenyl Pyrophosphate/Dimethylallyl Pyrophosphate Isomerase
J.Am.Chem.Soc., 127, 2005
|
|
2VNQ
 
 | | Monoclinic form of IDI-1 | | Descriptor: | IMIDAZOLE, ISOPENTENYL-DIPHOSPHATE DELTA-ISOMERASE, MANGANESE (II) ION, ... | | Authors: | de Ruyck, J, Oudjama, Y, Wouters, J. | | Deposit date: | 2008-02-06 | | Release date: | 2008-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Monoclinic Form of Isopentenyl Diphosphate Isomerase: A Case of Polymorphism in Biomolecular Crystals.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
7SP3
 
 | | E. coli RppH bound to Ap4A | | Descriptor: | BIS(ADENOSINE)-5'-TETRAPHOSPHATE, CHLORIDE ION, FLUORIDE ION, ... | | Authors: | Serganov, A.A, Vasilyev, N, Nuthanakanti, A. | | Deposit date: | 2021-11-02 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A distinct RNA recognition mechanism governs Np 4 decapping by RppH.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2W4E
 
 | | Structure of an N-terminally truncated Nudix hydrolase DR2204 from Deinococcus radiodurans | | Descriptor: | MUTT/NUDIX FAMILY PROTEIN | | Authors: | Goncalves, A.M.D, Fioravanti, E, Stelter, M, McSweeney, S. | | Deposit date: | 2008-11-25 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of an N-Terminally Truncated Nudix Hydrolase Dr2204 from Deinococcus Radiodurans.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
7T7H
 
 | | Crystal structure of Vaccinia Virus decapping enzyme D9 in complex with inhibitor CP100356 | | Descriptor: | 4-(6,7-dimethoxy-3,4-dihydroisoquinolin-2(1H)-yl)-N-[2-(3,4-dimethoxyphenyl)ethyl]-6,7-dimethoxyquinazolin-2-amine, DNA repair NTP-phosphohydrolase, SODIUM ION | | Authors: | Peters, J.K, Gross, J.D. | | Deposit date: | 2021-12-15 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.78000259 Å) | | Cite: | Fluorescence-Based Activity Screening Assay Reveals Small Molecule Inhibitors of Vaccinia Virus mRNA Decapping Enzyme D9.
Acs Chem.Biol., 17, 2022
|
|
1XSC
 
 | | Structure of the nudix enzyme AP4A hydrolase from homo sapiens (E63A mutant) in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Bis(5'-nucleosyl)-tetraphosphatase | | Authors: | Swarbrick, J.D, Buyya, S, Gunawardana, D, Gayler, K.R, McLennan, A.G, Gooley, P.R. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and Substrate-binding Mechanism of Human Ap4A Hydrolase
J.Biol.Chem., 280, 2005
|
|
1XSA
 
 | | Structure of the nudix enzyme AP4A hydrolase from homo sapiens (E63A mutant) | | Descriptor: | Bis(5'-nucleosyl)-tetraphosphatase | | Authors: | Swarbrick, J.D, Buyya, S, Gunawardana, D, Gayler, K.R, McLennan, A.G, Gooley, P.R. | | Deposit date: | 2004-10-18 | | Release date: | 2004-12-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and Substrate-binding Mechanism of Human Ap4A Hydrolase
J.Biol.Chem., 280, 2005
|
|
7TN4
 
 | | Diphosphoinositol polyphosphate phosphohydrolase 1 (DIPP1/NUDT3) in complex with 3-diphosphoinositol 1,2,4,5-tetrakisphosphate (3-PP-IP4), Mg and Fluoride ion | | Descriptor: | (1R,2S,3R,4R,5S,6S)-4-hydroxy-2,3,5,6-tetrakis(phosphonooxy)cyclohexyl trihydrogen diphosphate, CHLORIDE ION, Diphosphoinositol polyphosphate phosphohydrolase 1, ... | | Authors: | Zong, G, Wang, H, Shears, S.B. | | Deposit date: | 2022-01-20 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and catalytic analyses of the InsP 6 kinase activities of higher plant ITPKs.
Faseb J., 36, 2022
|
|
2VNP
 
 | | Monoclinic form of IDI-1 | | Descriptor: | 2-DIMETHYLAMINO-ETHYL-DIPHOSPHATE, ISOPENTENYL-DIPHOSPHATE DELTA-ISOMERASE, MAGNESIUM ION, ... | | Authors: | de Ruyck, J, Oudjama, Y, Wouters, J. | | Deposit date: | 2008-02-06 | | Release date: | 2008-03-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Monoclinic Form of Isopentenyl Diphosphate Isomerase: A Case of Polymorphism in Biomolecular Crystals.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
