3FFW
 
 | | Crystal Structure of CheY triple mutant F14Q, N59K, E89Y complexed with BeF3- and Mn2+ | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein cheY, GLYCEROL, ... | | Authors: | Pazy, Y, Collins, E.J, Bourret, R.B. | | Deposit date: | 2008-12-04 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Matching Biochemical Reaction Kinetics to the Timescales of Life: Structural Determinants That Influence the Autodephosphorylation Rate of Response Regulator Proteins.
J.Mol.Biol., 392, 2009
|
|
5CHY
 
 | | STRUCTURE OF CHEMOTAXIS PROTEIN CHEY | | Descriptor: | CALCIUM ION, CHEY | | Authors: | Zhu, X, Rebello, J, Matsumura, P, Volz, K. | | Deposit date: | 1996-08-29 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of CheY mutants Y106W and T87I/Y106W. CheY activation correlates with movement of residue 106.
J.Biol.Chem., 272, 1997
|
|
1E6K
 
 | | Two-component signal transduction system D12A mutant of CheY | | Descriptor: | Chemotaxis protein CheY | | Authors: | Sola, M, Lopez-Hernandez, E, Cronet, P, Lacroix, E, Serrano, L, Coll, M, Parraga, A. | | Deposit date: | 2000-08-18 | | Release date: | 2001-03-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Towards understanding a molecular switch mechanism: thermodynamic and crystallographic studies of the signal transduction protein CheY.
J.Mol.Biol., 303, 2000
|
|
3FFX
 
 | | Crystal Structure of CheY triple mutant F14E, N59R, E89H complexed with BeF3- and Mn2+ | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein cheY, GLYCEROL, ... | | Authors: | Pazy, Y, Collins, E.J, Bourret, R.B. | | Deposit date: | 2008-12-04 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Matching Biochemical Reaction Kinetics to the Timescales of Life: Structural Determinants That Influence the Autodephosphorylation Rate of Response Regulator Proteins.
J.Mol.Biol., 392, 2009
|
|
3OLY
 
 | |
1VLZ
 
 | |
3RVS
 
 | | Structure of the CheYN59D/E89R Tungstate complex | | Descriptor: | Chemotaxis protein CheY, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Immormino, R.M, Starbird, C.A, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3RVR
 
 | | Structure of the CheYN59D/E89R Molybdate complex | | Descriptor: | Chemotaxis protein CheY, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Immormino, R.M, Starbird, C.A, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
3MYY
 
 | |
3RVJ
 
 | | Structure of the CheY-BeF3 Complex with substitutions at 59 and 89: N59D and E89Q | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein CheY, GLYCEROL, ... | | Authors: | Starbird, C.A, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
1FFG
 
 | | CHEY-BINDING DOMAIN OF CHEA IN COMPLEX WITH CHEY AT 2.1 A RESOLUTION | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, CHEMOTAXIS PROTEIN CHEY, MANGANESE (II) ION | | Authors: | Gouet, P, Chinardet, N, Welch, M, Guillet, V, Birck, C, Mourey, L, Samama, J.-P. | | Deposit date: | 2000-07-25 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Further insights into the mechanism of function of the response regulator CheY from crystallographic studies of the CheY--CheA(124--257) complex.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
3OLX
 
 | |
1AB6
 
 | | STRUCTURE OF CHEY MUTANT F14N, V86T | | Descriptor: | CHEMOTAXIS PROTEIN CHEY | | Authors: | Wilcock, D, Pisabarro, M.T, Lopez-Hernandez, E, Serranno, L, Coll, M. | | Deposit date: | 1997-02-04 | | Release date: | 1998-02-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of two CheY mutants: importance of the hydrogen-bond contribution to protein stability.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
3FFT
 
 | | Crystal Structure of CheY double mutant F14E, E89R complexed with BeF3- and Mn2+ | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein cheY, GLYCEROL, ... | | Authors: | Pazy, Y, Collins, E.J, Bourret, R.B. | | Deposit date: | 2008-12-04 | | Release date: | 2009-09-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Matching Biochemical Reaction Kinetics to the Timescales of Life: Structural Determinants That Influence the Autodephosphorylation Rate of Response Regulator Proteins.
J.Mol.Biol., 392, 2009
|
|
1F4V
 
 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY BOUND TO THE N-TERMINUS OF FLIM | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, FLAGELLAR MOTOR SWITCH PROTEIN, ... | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Henderson, R.K, King, D, Huang, L.S, Kustu, S, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-06-10 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of an activated response regulator bound to its target.
Nat.Struct.Biol., 8, 2001
|
|
1HEY
 
 | |
3RVN
 
 | | Structure of the CheY-BeF3 Complex with substitutions at 59 and 89: N59D and E89Y | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Chemotaxis protein CheY, GLYCEROL, ... | | Authors: | Starbird, C.A, Immormino, R.M, Silversmith, R.E, Bourret, R.B. | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Probing Mechanistic Similarities between Response Regulator Signaling Proteins and Haloacid Dehalogenase Phosphatases.
Biochemistry, 54, 2015
|
|
1EHC
 
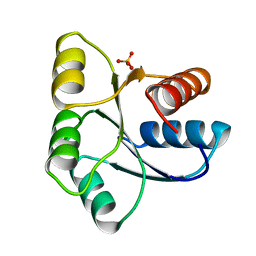 | | STRUCTURE OF SIGNAL TRANSDUCTION PROTEIN CHEY | | Descriptor: | CHEY, SULFATE ION | | Authors: | Jiang, M, Bourret, R, Simon, M, Volz, K. | | Deposit date: | 1996-03-05 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Uncoupled phosphorylation and activation in bacterial chemotaxis. The 2.3 A structure of an aspartate to lysine mutant at position 13 of CheY.
J.Biol.Chem., 272, 1997
|
|
1YMU
 
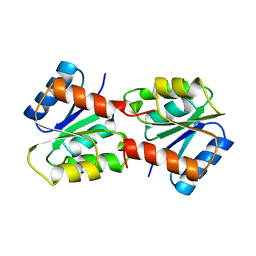 | |
3OLW
 
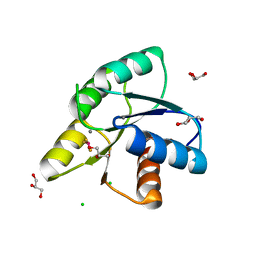 | |
6CHY
 
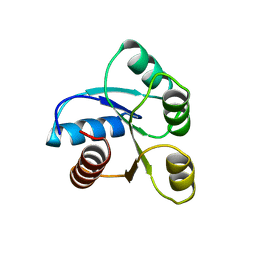 | | STRUCTURE OF CHEMOTAXIS PROTEIN CHEY | | Descriptor: | CHEY, SULFATE ION | | Authors: | Zhu, X, Rebello, J, Matsumura, P, Volz, K. | | Deposit date: | 1996-08-29 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Crystal structures of CheY mutants Y106W and T87I/Y106W. CheY activation correlates with movement of residue 106.
J.Biol.Chem., 272, 1997
|
|
1FQW
 
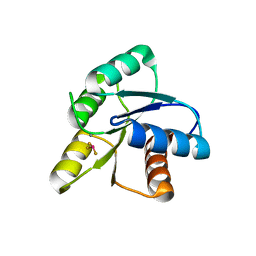 | | CRYSTAL STRUCTURE OF ACTIVATED CHEY | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CHEMOTAXIS CHEY PROTEIN, MANGANESE (II) ION | | Authors: | Lee, S.Y, Cho, H.S, Pelton, J.G, Yan, D, Berry, E.A, Wemmer, D.E. | | Deposit date: | 2000-09-07 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of activated CheY. Comparison with other activated receiver domains.
J.Biol.Chem., 276, 2001
|
|
1FFS
 
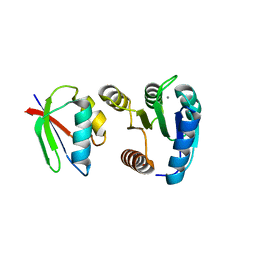 | | CHEY-BINDING DOMAIN OF CHEA IN COMPLEX WITH CHEY FROM CRYSTALS SOAKED IN ACETYL PHOSPHATE | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, CHEMOTAXIS PROTEIN CHEY, MANGANESE (II) ION | | Authors: | Gouet, P, Chinardet, N, Welch, M, Guillet, V, Birck, C, Mourey, L, Samama, J.-P. | | Deposit date: | 2000-07-26 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Further insights into the mechanism of function of the response regulator CheY from crystallographic studies of the CheY--CheA(124--257) complex.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1AB5
 
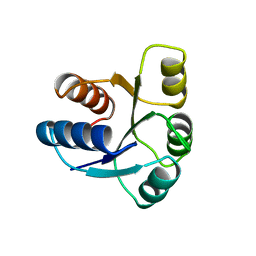 | | STRUCTURE OF CHEY MUTANT F14N, V21T | | Descriptor: | CHEY | | Authors: | Wilcock, D, Pisabarro, M.T, Lopez-Hernandez, E, Serrano, L, Coll, M. | | Deposit date: | 1997-02-04 | | Release date: | 1998-02-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure analysis of two CheY mutants: importance of the hydrogen-bond contribution to protein stability.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1ZDM
 
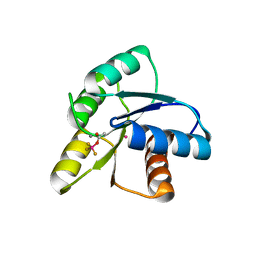 | | Crystal Structure of Activated CheY Bound to Xe | | Descriptor: | Chemotaxis protein cheY, MANGANESE (II) ION, XENON | | Authors: | Lowery, T.J, Doucleff, M, Ruiz, E.J, Rubin, S.M, Pines, A, Wemmer, D.E. | | Deposit date: | 2005-04-14 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Distinguishing multiple chemotaxis Y protein conformations with laser-polarized 129Xe NMR.
Protein Sci., 14, 2005
|
|
